6HTJ
 
 | |
8BWT
 
 | |
2K3F
 
 | |
8PE0
 
 | | X-ray structure of the Thermus thermophilus K167L mutant of the PilF-GSPIIB domain in the c-di-GMP bound state | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, SULFATE ION, ... | | Authors: | Neissner, K, Woehnert, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the K167L mutant of PilF-GSPIIB domain in the c-di-GMP bound state
To Be Published
|
|
8PDK
 
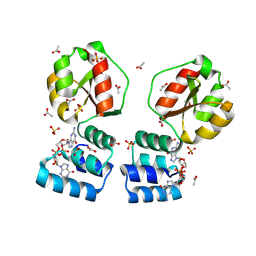 | | X-ray structure of the Thermus thermophilus PilF-GSPIIB domain in the c-di-GMP bound state | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, SULFATE ION, ... | | Authors: | Neissner, K, Woehnert, J. | | Deposit date: | 2023-06-12 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the PilF-GSPIIB domain in the c-di-GMP bound state
To Be Published
|
|
8PFA
 
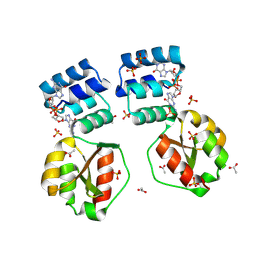 | | X-ray structure of the Thermus thermophilus K167R mutant of the PilF-GSPIIB domain in the c-di-GMP bound state | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, SULFATE ION, ... | | Authors: | Neissner, K, Woehnert, J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the Thermus thermophilus K167R mutant of the PilF-GSPIIB domain in the c-di-GMP bound state
To Be Published
|
|
8PKZ
 
 | |
8PQU
 
 | |
6TRP
 
 | | Solution Structure of Docking Domain Complex of Pax NRPS: PaxC NDD - PaxB CDD | | Descriptor: | Peptide synthetase XpsB,Peptide synthetase XpsB | | Authors: | Watzel, J, Hacker, C, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A New Docking Domain Type in the Peptide-Antimicrobial-Xenorhabdus Peptide Producing Nonribosomal Peptide Synthetase fromXenorhabdus bovienii.
Acs Chem.Biol., 15, 2020
|
|
2JQ7
 
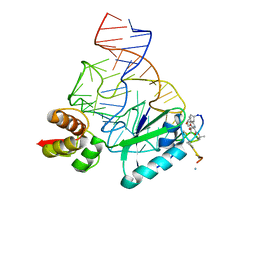 | | Model for thiostrepton binding to the ribosomal L11-RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RIBOSOMAL RNA, THIOSTREPTON | | Authors: | Jonker, H.R.A, Ilin, S, Grimm, S.K, Woehnert, J, Schwalbe, H. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | L11 Domain Rearrangement Upon Binding to RNA and Thiostrepton Studied by NMR Spectroscopy
Nucleic Acids Res., 35, 2007
|
|
2MZJ
 
 | |
3RKF
 
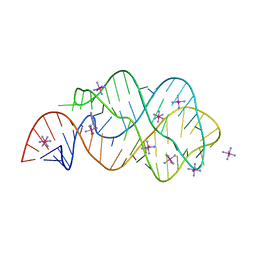 | | Crystal structure of guanine riboswitch C61U/G37A double mutant bound to thio-guanine | | Descriptor: | 2-amino-1,9-dihydro-6H-purine-6-thione, COBALT HEXAMMINE(III), Guanine riboswitch | | Authors: | Buck, J, Wacker, A, Warkentin, E, Woehnert, J, Wirmer-Bartoschek, J, Schwalbe, H. | | Deposit date: | 2011-04-18 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Influence of ground-state structure and Mg2+ binding on folding kinetics of the guanine-sensing riboswitch aptamer domain.
Nucleic Acids Res., 39, 2011
|
|
5LWJ
 
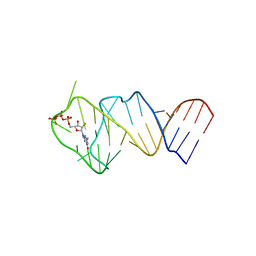 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6S10
 
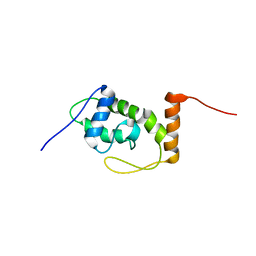 | |
1B75
 
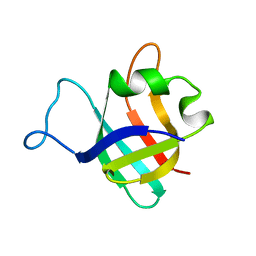 | |
5APG
 
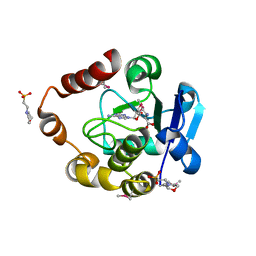 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
5AP8
 
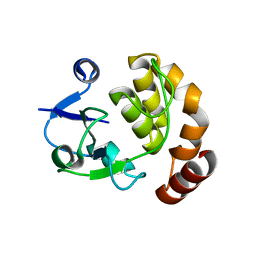 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | Descriptor: | TSR3 | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
7B2B
 
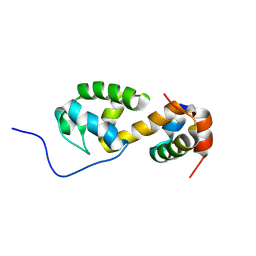 | | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | | Descriptor: | Amino acid adenylation domain-containing protein, Peptide synthetase PaxA | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B2F
 
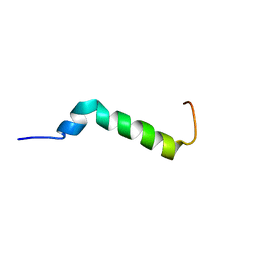 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6EWU
 
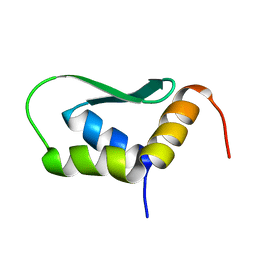 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWS
 
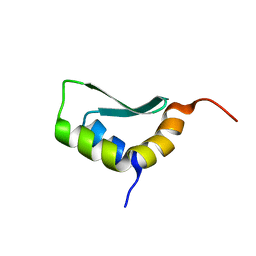 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWV
 
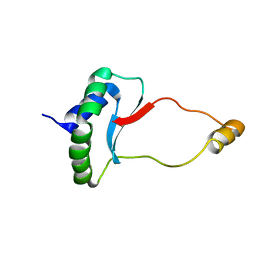 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | Descriptor: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWT
 
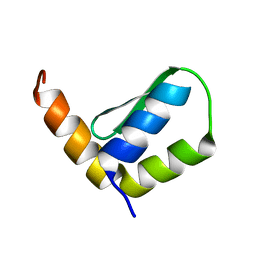 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
2N0J
 
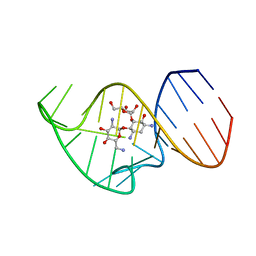 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | Descriptor: | RIBOSTAMYCIN, RNA_(27-MER) | | Authors: | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | Deposit date: | 2015-03-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2KMJ
 
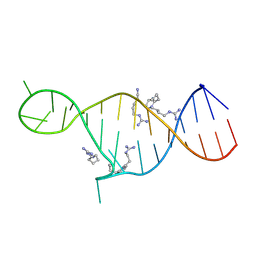 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
