3LZM
 
 | |
149L
 
 | |
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2021-06-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
3ZSE
 
 | | 3D Structure of a thermophilic family GH11 xylanase from Thermobifida fusca | | 分子名称: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | 著者 | Lammerts van Bueren, A, Otani, S, Friis, E.P, S Wilson, K, Davies, G.J. | | 登録日 | 2011-06-27 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Three-Dimensional Structure of a Thermophilic Family Gh11 Xylanase from Thermobifida Fusca.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2021-06-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5FN0
 
 | | Crystal structure of Pseudomonas fluorescens kynurenine-3- monooxygenase (KMO) in complex with GSK180 | | 分子名称: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, KYNURENINE 3-MONOOXYGENASE | | 著者 | Mole, D.J, Webster, S.P, Uings, I, Zheng, X, Binnie, M, Wilson, K, Hutchinson, J.P, Mirguet, O, Walker, A, Beaufils, B, Ancellin, N, Trottet, L, Beneton, V, Mowat, C.G, Wilkinson, M, Rowland, P, Haslam, C, McBride, A, Homer, N.Z.M, Baily, J.E, Sharp, M.G.F, Garden, O.J, Hughes, J, Howie, S.E.M, Holmes, D, Liddle, J, Iredale, J.P. | | 登録日 | 2015-11-10 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Kynurenine-3-Monooxygenase Inhibition Prevents Multiple Organ Failure in Rodent Models of Acute Pancreatitis.
Nat.Med. (N.Y.), 22, 2016
|
|
5NTG
 
 | |
5NTF
 
 | |
5NSM
 
 | |
5NTH
 
 | |
5NSQ
 
 | |
5NTD
 
 | | Structure of Leucyl aminopeptidase from Trypanosoma brucei in complex with Bestatin | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BICARBONATE ION, ... | | 著者 | Timm, J, Wilson, K. | | 登録日 | 2017-04-27 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | 分子名称: | CHLORIDE ION, T4 LYSOZYME | | 著者 | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | 登録日 | 1991-01-25 | | 公開日 | 1992-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8RXN
 
 | | REFINEMENT OF RUBREDOXIN FROM DESULFOVIBRIO VULGARIS AT 1.0 ANGSTROMS WITH AND WITHOUT RESTRAINTS | | 分子名称: | FE (III) ION, RUBREDOXIN, SULFATE ION | | 著者 | Dauter, Z, Sieker, L, Wilson, K. | | 登録日 | 1991-08-26 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Refinement of rubredoxin from Desulfovibrio vulgaris at 1.0 A with and without restraints.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1OBG
 
 | | SAICAR-synthase complexed with ATP | | 分子名称: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHORIBOSYLAMIDOIMIDAZOLE- SUCCINOCARBOXAMIDE SYNTHASE, ... | | 著者 | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | 登録日 | 2003-01-30 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBD
 
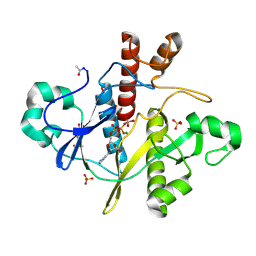 | | SAICAR-synthase complexed with ATP | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | 登録日 | 2003-01-30 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
3BX1
 
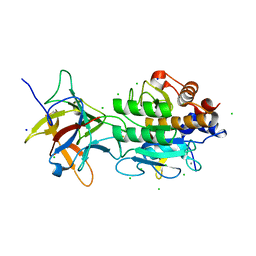 | | Complex between the Barley alpha-Amylase/Subtilisin Inhibitor and the subtilisin Savinase | | 分子名称: | Alpha-amylase/subtilisin inhibitor, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Micheelsen, P.O, Vevodova, J, Wilson, K, Skjot, M. | | 登録日 | 2008-01-11 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Mutational Analyses of the Interaction between the Barley alpha-Amylase/Subtilisin Inhibitor and the Subtilisin Savinase Reveal a Novel Mode of Inhibition
J.Mol.Biol., 380, 2008
|
|
3CYR
 
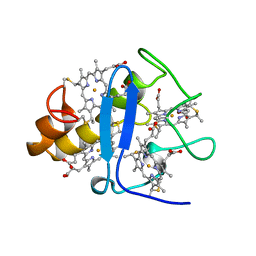 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774P | | 分子名称: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | 登録日 | 1997-07-24 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Refinement of the Three-Dimensional Structures of Cytochrome C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstroms Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstroms Resolution
Inorg.Chim.Acta., 273, 1998
|
|
2CTH
 
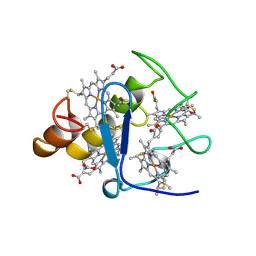 | | CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH | | 分子名称: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | 登録日 | 1997-06-18 | | 公開日 | 1997-12-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Refinement of the Three-Dimensional Structures of Cytochromes C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstrom Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstrom Resolution
Inorg.Chim.Acta., 273, 1998
|
|
1N52
 
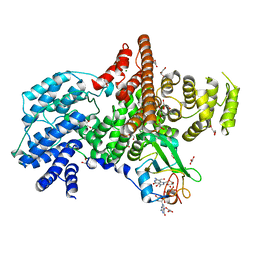 | | Cap Binding Complex | | 分子名称: | 20 kDa nuclear cap binding protein, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 80 kDa nuclear cap binding protein, ... | | 著者 | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | 登録日 | 2002-11-04 | | 公開日 | 2003-02-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1N54
 
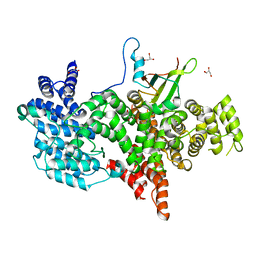 | | Cap Binding Complex m7GpppG free | | 分子名称: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | 著者 | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | 登録日 | 2002-11-04 | | 公開日 | 2003-02-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1L06
 
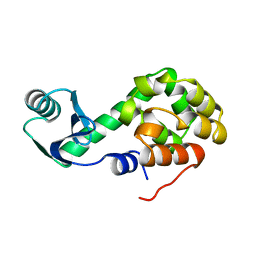 | |
1L03
 
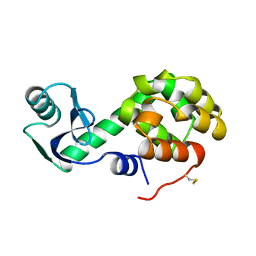 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | 著者 | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | 登録日 | 1988-02-05 | | 公開日 | 1988-04-16 | | 最終更新日 | 2022-11-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
1L11
 
 | | CONTRIBUTIONS OF HYDROGEN BONDS OF THR 157 TO THE THERMODYNAMIC STABILITY OF PHAGE T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | 著者 | Dao-Pin, S, Wilson, K, Alber, T, Matthews, B.W. | | 登録日 | 1988-02-05 | | 公開日 | 1988-04-16 | | 最終更新日 | 2022-11-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme.
Nature, 330, 1987
|
|
