2L6Z
 
 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID with FOG | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L6Y
 
 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3VEK
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
3VD6
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P21 Crystal Form | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | Descriptor: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | Authors: | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
6BGG
 
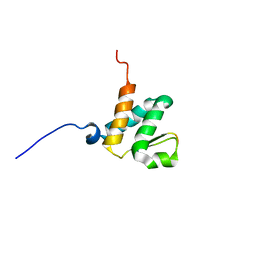 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
6CVD
 
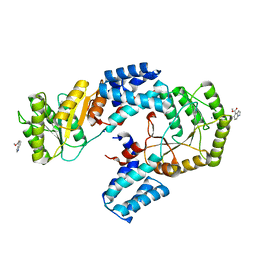 | | High resolution crystal structure of FtsY-NG domain of E. coli bound to fragment 1 | | Descriptor: | AMMONIUM ION, SODIUM ION, Signal recognition particle receptor FtsY, ... | | Authors: | Faoro, C, Ataide, S.F, Kwan, A, Wilkinson-White, L. | | Deposit date: | 2018-03-27 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of fragments that target key interactions in the signal recognition particle (SRP) as potential leads for a new class of antibiotics.
PLoS ONE, 13, 2018
|
|
