2PFM
 
 | | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis | | Descriptor: | Adenylosuccinate lyase, MALONATE ION | | Authors: | Levdikov, V.M, Blagova, E.V, Baumgart, M, Moroz, O.V, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis
To be Published
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4X
 
 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4Y
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
2TGF
 
 | | THE SOLUTION STRUCTURE OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA | | Descriptor: | TRANSFORMING GROWTH FACTOR-ALPHA | | Authors: | Harvey, T.S, Wilkinson, A.J, Tappin, M.J, Cooke, R.M, Campbell, I.D. | | Deposit date: | 1991-01-23 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human transforming growth factor alpha.
Eur.J.Biochem., 198, 1991
|
|
2V36
 
 | | Crystal structure of gamma-glutamyl transferase from Bacillus subtilis | | Descriptor: | GAMMA-GLUTAMYLTRANSPEPTIDASE LARGE CHAIN, GAMMA-GLUTAMYLTRANSPEPTIDASE SMALL CHAIN | | Authors: | Sharath, B, Prabhune, A.A, Suresh, C.G, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2007-06-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Gamma-Glutamyl Transferase
To be Published
|
|
2V25
 
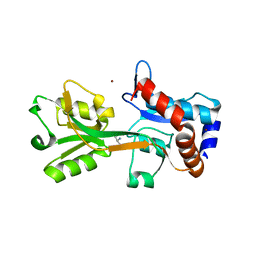 | | Structure of the Campylobacter jejuni antigen Peb1A, an aspartate and glutamate receptor with bound aspartate | | Descriptor: | ASPARTIC ACID, MAJOR CELL-BINDING FACTOR, ZINC ION | | Authors: | Muller, A, Dodson, E, del Rocio Leon-Kempis, M, Kelly, D.J, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Bacterial Virulence Factor with a Dual Role as an Adhesin and a Solute Binding-Protein: The Crystal Structure at 1.5 A Resolution of the Peb1A Protein from the Food-Borne Human Pathogen Campylobacter Jejuni
J.Mol.Biol., 372, 2007
|
|
2WCE
 
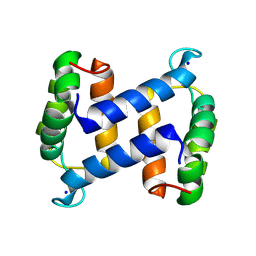 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCF
 
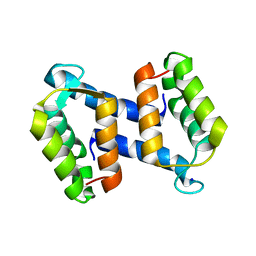 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
1LV7
 
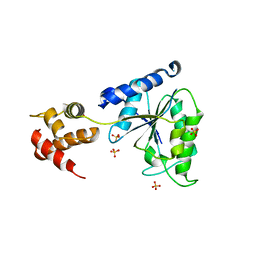 | | Crystal Structure of the AAA domain of FtsH | | Descriptor: | FtsH, SULFATE ION | | Authors: | Krzywda, S, Brzozowski, A.M, Verma, C, Karata, K, Ogura, T, Wilkinson, A.J. | | Deposit date: | 2002-05-26 | | Release date: | 2002-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution.
Structure, 10, 2002
|
|
1XOC
 
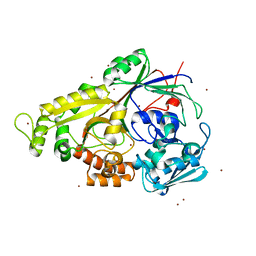 | | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. | | Descriptor: | Nonapeptide VDSKNTSSW, Oligopeptide-binding protein appA, ZINC ION | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wright, L, Vagin, A.A, Wilkinson, A.J. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide.
J.Mol.Biol., 345, 2005
|
|
8AY0
 
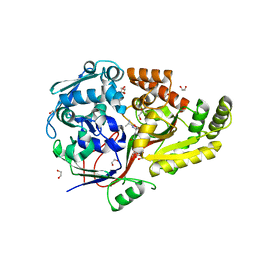 | | Crystal Structure of the peptide binding protein DppE from Bacillus subtilis in complex with murein tripeptide | | Descriptor: | 1,2-ETHANEDIOL, Dipeptide-binding protein DppE, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Hughes, A.M, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8AZB
 
 | |
5DOL
 
 | | Crystal structure of YabA amino-terminal domain from Bacillus subtilis | | Descriptor: | Initiation-control protein YabA | | Authors: | Cherrier, M.V, Bazin, A, Jameson, K.H, Wilkinson, A.J, Noirot-Gros, M.F, Terradot, L. | | Deposit date: | 2015-09-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners.
Nucleic Acids Res., 44, 2016
|
|
5G21
 
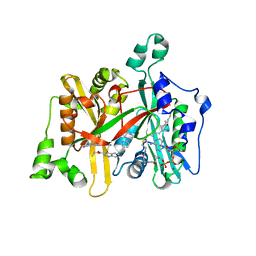 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G1Z
 
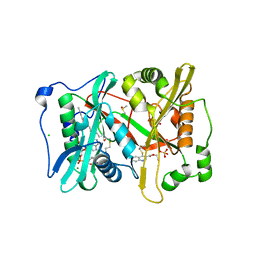 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 1) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G20
 
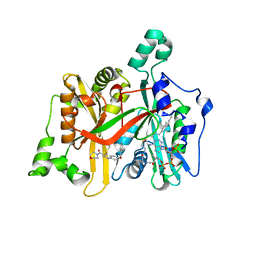 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 19). | | Descriptor: | 6-(BENZYLOXY)-4-(ETHYLSULFANYL)-3-[(MORPHOLIN-4-YL), DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
1QMP
 
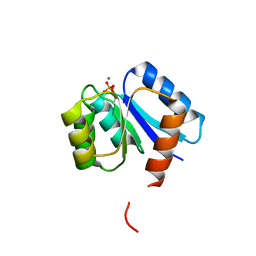 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
5G22
 
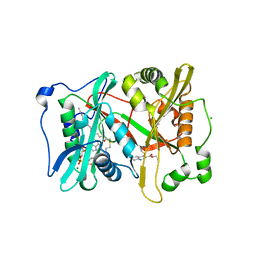 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 26) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
8ARE
 
 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with a PhrE-derived pentapeptide | | Descriptor: | Oligopeptide-binding protein OppA, Phosphatase RapE inhibitor, SULFATE ION | | Authors: | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8ARN
 
 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with an endogenous tetrapeptide | | Descriptor: | Endogenous tetrapeptide (SER-ASN-SER-SER), Oligopeptide-binding protein OppA | | Authors: | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
2A1Y
 
 | | Crystal Structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A Resolution. | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of GuaC-GMP complex from Bacillus anthracis at 2.26 A resolution.
To be Published
|
|
2B18
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-15 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
2B0L
 
 | | C-terminal DNA binding domain of transcriptional pleiotropic repressor CodY. | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor codY | | Authors: | Levdikov, V.M, Blagova, E, Joseph, P, Sonenshein, A.L, Wilkinson, A.J. | | Deposit date: | 2005-09-14 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of CodY, a GTP- and Isoleucine-responsive Regulator of Stationary Phase and Virulence in Gram-positive Bacteria.
J.Biol.Chem., 281, 2006
|
|
2BPI
 
 | | Structure of Iron dependent superoxide dismutase from P. falciparum. | | Descriptor: | FE (III) ION, FE-SUPEROXIDE DISMUTASE | | Authors: | Boucher, I.W, Brannigan, J, Wilkinson, A.J, Brzozowski, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Crystal Structure of Superoxide Dismutase from Plasmodium Falciparum.
Bmc Struct.Biol., 6, 2006
|
|
