4V41
 
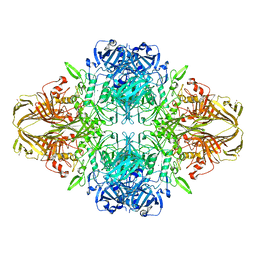 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2014-07-09 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
5DMA
 
 | |
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | 著者 | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | 登録日 | 2017-06-21 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.06 Å) | | 主引用文献 | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3U4Q
 
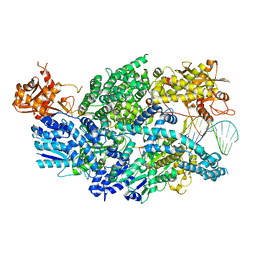 | | Structure of AddAB-DNA complex at 2.8 angstroms | | 分子名称: | 1,2-ETHANEDIOL, ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, ... | | 著者 | Saikrishnan, K, Krajewski, W, Wigley, D. | | 登録日 | 2011-10-10 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
3U44
 
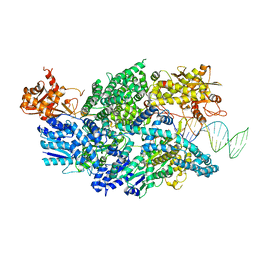 | | Crystal structure of AddAB-DNA complex | | 分子名称: | ATP-dependent helicase/deoxyribonuclease subunit B, ATP-dependent helicase/nuclease subunit A, DNA (36-MER), ... | | 著者 | Saikrishnan, K, Krajewski, W, Wigley, D. | | 登録日 | 2011-10-07 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex.
Embo J., 31, 2012
|
|
1DP0
 
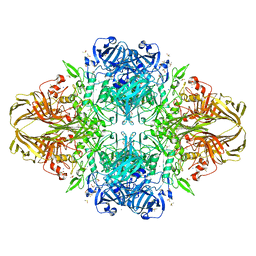 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | 分子名称: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 1999-12-22 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
6FWS
 
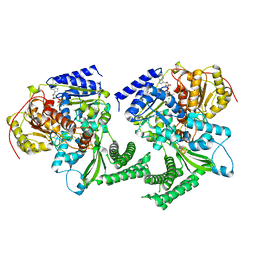 | |
4CEH
 
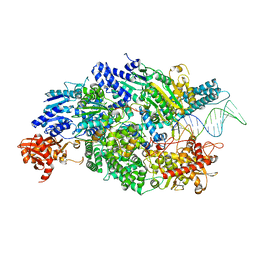 | | Crystal structure of AddAB with a forked DNA substrate | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4CEI
 
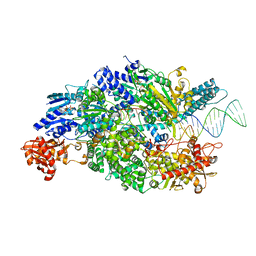 | | Crystal structure of ADPNP-bound AddAB with a forked DNA substrate | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4CEJ
 
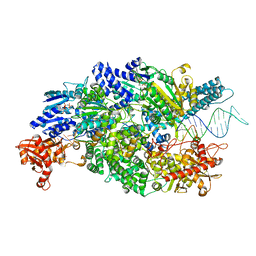 | | Crystal structure of AddAB-DNA-ADPNP complex at 3 Angstrom resolution | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
1F4H
 
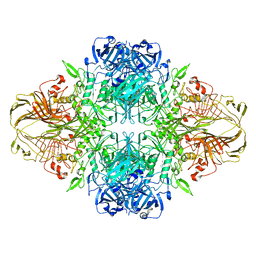 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
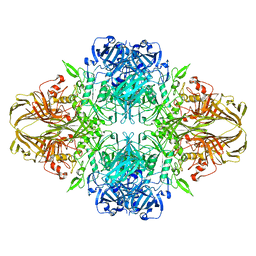 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | 分子名称: | BETA-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 2000-06-07 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
