2ML8
 
 | |
2LFB
 
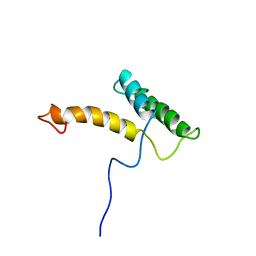 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | LFB1/HNF1 TRANSCRIPTION FACTOR | | Authors: | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | Deposit date: | 1996-12-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
2L4F
 
 | |
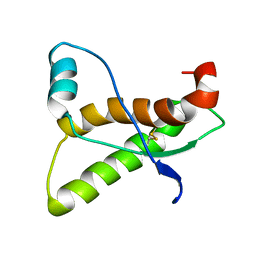
3CYS
 
 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF THE CYCLOPHILIN A-CYCLOSPORIN A COMPLEX | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Spitzfaden, C, Braun, W, Wider, G, Widmer, H, Wuthrich, K. | | Deposit date: | 1994-02-28 | | Release date: | 1994-08-31 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR Solution Structure of the Cyclophilin A-Cyclosporin a Complex.
J.Biomol.NMR, 4, 1994
|
|
1AG2
 
 | | PRION PROTEIN DOMAIN PRP(121-231) FROM MOUSE, NMR, 2 MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Billeter, M, Riek, R, Wider, G, Wuthrich, K, Hornemann, S, Glockshuber, R. | | Deposit date: | 1997-03-31 | | Release date: | 1997-10-08 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mouse prion protein domain PrP(121-231).
Nature, 382, 1996
|
|
1KX6
 
 | |
1SQ8
 
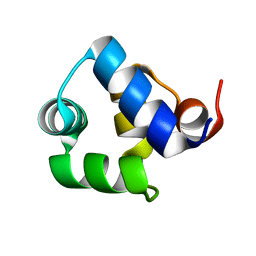 | |
1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
2MN5
 
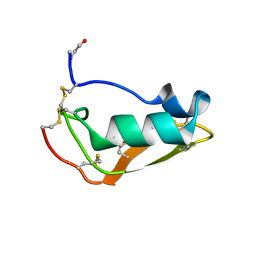 | | NMR structure of Copsin | | Descriptor: | Copsin | | Authors: | Hofmann, D, Wider, G, Essig, A, Aebi, M. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Copsin, a Novel Peptide-based Fungal Antibiotic Interfering with the Peptidoglycan Synthesis.
J.Biol.Chem., 289, 2014
|
|
2KWV
 
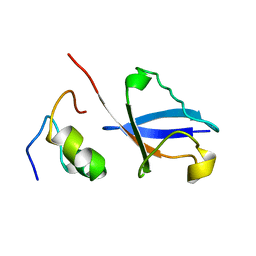 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWU
 
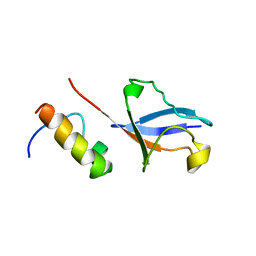 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
1PBA
 
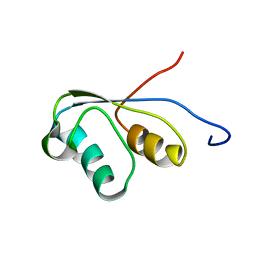 | | THE NMR STRUCTURE OF THE ACTIVATION DOMAIN ISOLATED FROM PORCINE PROCARBOXYPEPTIDASE B | | Descriptor: | PROCARBOXYPEPTIDASE B | | Authors: | Vendrell, J, Wider, G, Billeter, M, Aviles, F.X, Wuthrich, K. | | Deposit date: | 1991-11-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the activation domain isolated from porcine procarboxypeptidase B.
EMBO J., 10, 1991
|
|
2L4E
 
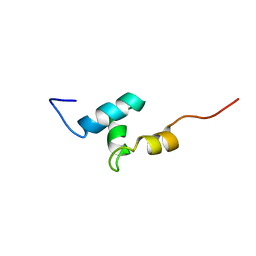 | |
2M5G
 
 | | Solution structure of FimA wt | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Walczak, M.J, Puorger, C, Glockshuber, R, Wider, G. | | Deposit date: | 2013-02-24 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Intramolecular donor strand complementation in the E. coli type 1 pilus subunit FimA explains the existence of FimA monomers as off-pathway products of pilus assembly that inhibit host cell apoptosis.
J.Mol.Biol., 426, 2014
|
|
2JTY
 
 | | Self-complemented variant of FimA, the main subunit of type 1 pilus | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Erilov, D, Wider, G, Glockshuber, R, Puorger, C, Vetsch, M. | | Deposit date: | 2007-08-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structure, Folding and Stability of FimA, the Main Structural Subunit of Type 1 Pili from Uropathogenic Escherichia coli Strains.
J.Mol.Biol., 412, 2011
|
|
1PN5
 
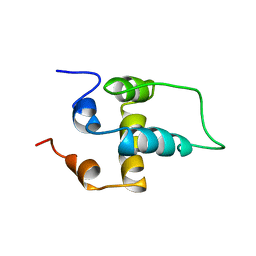 | | NMR structure of the NALP1 Pyrin domain (PYD) | | Descriptor: | NACHT-, LRR- and PYD-containing protein 2 | | Authors: | Hiller, S, Kohl, A, Fiorito, F, Herrmann, T, Wider, G, Tschopp, J, Grutter, M.G, Wuthrich, K. | | Deposit date: | 2003-06-12 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain
Structure, 11, 2003
|
|
1QJL
 
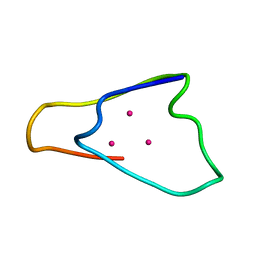 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1QJK
 
 | | Metallothionein MTA from sea urchin (alpha domain) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
