5OV2
 
 | |
7ZEM
 
 | |
7ZEO
 
 | |
7ZEK
 
 | |
8ABD
 
 | |
8ABN
 
 | |
2VCD
 
 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
2KY7
 
 | | NMR Structural Studies on the Covalent DNA Binding of a Pyrrolobenzodiazepine-Naphthalimide Conjugate | | Descriptor: | 2-{2-[4-(3-{[(11aS)-7-methoxy-5-oxo-2,3,5,10,11,11a-hexahydro-1H-pyrrolo[2,1-c][1,4]benzodiazepin-8-yl]oxy}propyl)piperazin-1-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Langel, W, Kamal, A, Weisz, K. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate
Org.Biomol.Chem., 8, 2010
|
|
2LQ8
 
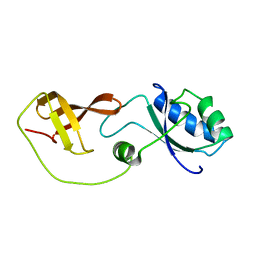 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
2LPX
 
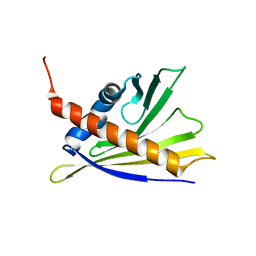 | | Solution Structure of Strawberry Allergen Fra a 1e | | Descriptor: | Major strawberry allergen Fra a 1-E | | Authors: | Seutter von Loetzen, C, Hartl-Spiegelhauer, O, Schweimer, K, Schwab, W, Roesch, P. | | Deposit date: | 2012-02-20 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the strawberry allergen Fra a 1.
Biosci.Rep., 32, 2012
|
|
2LSN
 
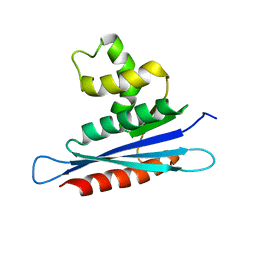 | | Solution structure of PFV RNase H domain | | Descriptor: | RNase H | | Authors: | Leo, B, Schweimer, K, Woehrl, B. | | Deposit date: | 2012-05-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the prototype foamy virus RNase H domain indicates an important role of the basic loop in substrate binding.
Retrovirology, 9, 2012
|
|
2MEW
 
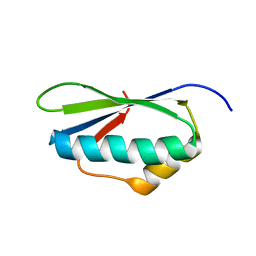 | |
2JX2
 
 | | Solution conformation of RNA-bound NELF-E RRM | | Descriptor: | Negative elongation factor E | | Authors: | Jampani, N, Schweimer, K, Wenzel, S, Woehrl, B.M, Roesch, P. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NELF-E RRM Undergoes Major Structural Changes in Flexible Protein Regions on Target RNA Binding
Biochemistry, 47, 2008
|
|
2N3E
 
 | | Amino-terminal domain of Latrodectus hesperus MaSp1 with neutralized acidic cluster | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Schaal, D, Bauer, J, Schweimer, K, Scheibel, T, Roesch, P, Schwarzinger, S. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of an engineered amino-terminal ampullate spider silk with neutralized charge cluster
To be Published
|
|
2JYS
 
 | |
2JZB
 
 | |
2LCL
 
 | |
2K7H
 
 | | NMR solution structure of soybean allergen Gly m 4 | | Descriptor: | Stress-induced protein SAM22 | | Authors: | Berkner, H, Neudecker, P, Mittag, D, Ballmer-Weber, B.K, Schweimer, K, Vieths, S, Roesch, P. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins
Biosci.Rep., 29, 2009
|
|
2KVQ
 
 | |
2MI6
 
 | |
