1SY0
 
 | | 1.15 A Crystal Structure of T121V Mutant of Nitrophorin 4 from Rhodnius Prolixus | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1NP4
 
 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, PROTEIN (NITROPHORIN 4), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Weichsel, A, Champagne, D.E, Balfour, C.A, Montfort, W.R. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein.
Structure, 6, 1998
|
|
1SXU
 
 | | 1.4 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Imidazole | | Descriptor: | IMIDAZOLE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SY3
 
 | | 1.00 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1R8C
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | MANGANESE (II) ION, SODIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1SXW
 
 | | 1.05 A Crystal Structure of D30A Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1SY2
 
 | | 1.0 A Crystal Structure of D129A/L130A Mutant of Nitrophorin 4 | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1R89
 
 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1SXY
 
 | | 1.07 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1RO0
 
 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1- Triple mutant F50M/L107M/L110M SeMet remote | | Descriptor: | ORF904, ZINC ION | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1FZI
 
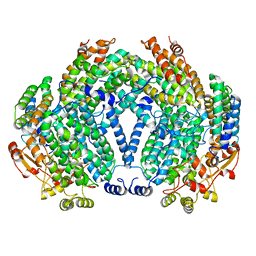 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM I PRESSURIZED WITH XENON GAS | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ALPHA CHAIN, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1FZ9
 
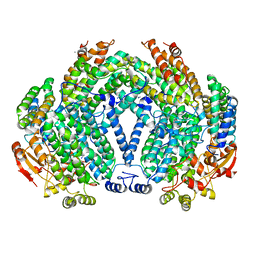 | | Methane monooxygenase hydroxylase, form II cocrystallized with iodoethane | | Descriptor: | CALCIUM ION, FE (III) ION, IODOETHANE, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
1R8A
 
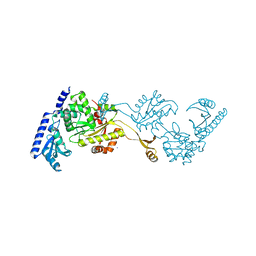 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | MANGANESE (II) ION, SODIUM ION, tRNA nucleotidyltransferase | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R65
 
 | | Crystal structure of ferrous soaked Ribonucleotide Reductase R2 subunit (wildtype) at pH 5 from E. coli | | Descriptor: | FE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1R8B
 
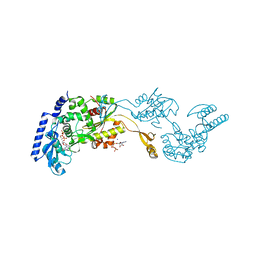 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1RO2
 
 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1- Triple mutant F50M/L107M/L110M manganese soak | | Descriptor: | MANGANESE (II) ION, ZINC ION, hypothetical protein ORF904 | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
4FXM
 
 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
1IKE
 
 | | Crystal Structure of Nitrophorin 4 from Rhodnius Prolixus Complexed with Histamine at 1.5 A Resolution | | Descriptor: | HISTAMINE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qiu, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1F5L
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN-AMILORIDE COMPLEX | | Descriptor: | 3,5-DIAMINO-N-(AMINOIMINOMETHYL)-6-CHLOROPYRAZINECARBOXAMIDE, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Zeslawska, E, Schweinitz, A, Karcher, A, Sondermann, P, Sperl, S, Sturzebecher, J, Jacob, U. | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals of the urokinase type plasminogen activator variant beta(c)-uPAin complex with small molecule inhibitors open the way towards structure-based drug design.
J.Mol.Biol., 301, 2000
|
|
1U96
 
 | | Solution Structure of Yeast Cox17 with Copper Bound | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Abajian, C, Yatsunyk, L.A, Ramirez, B.E, Rosenzweig, A.C. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Yeast cox17 solution structure and Copper(I) binding.
J.Biol.Chem., 279, 2004
|
|
4CR2
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1U97
 
 | |
4CR4
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CR3
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1G5B
 
 | | BACTERIOPHAGE LAMBDA SER/THR PROTEIN PHOSPHATASE | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE, ... | | Authors: | Voegtli, W.C, White, D.J, Reiter, N.J, Rusnak, F, Rosenzweig, A.C. | | Deposit date: | 2000-10-31 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the bacteriophage lambda Ser/Thr protein phosphatase with sulfate ion bound in two coordination modes.
Biochemistry, 39, 2000
|
|
