6DAM
 
 | | Crystal structure of lanthanide-dependent methanol dehydrogenase XoxF from Methylomicrobium buryatense 5G | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Deng, Y, Ro, S.Y, Rosenzweig, A.C. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C.
J. Biol. Inorg. Chem., 23, 2018
|
|
8SR2
 
 | | particulate methane monooxygenase incubated with 4,4,4-trifluorobutanol | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Product analogue binding identifies the copper active site of particulate methane monooyxgenase
Nat Catal, 2023
|
|
5D1M
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (P199A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
6FLQ
 
 | |
6HAP
 
 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
8OYI
 
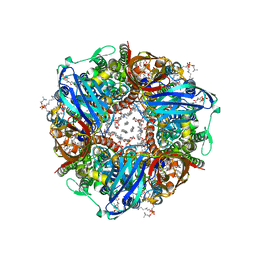 | | particulate methane monooxygenase with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
6HAM
 
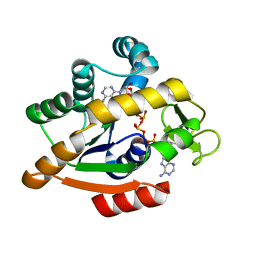 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
6CXH
 
 | |
5C91
 
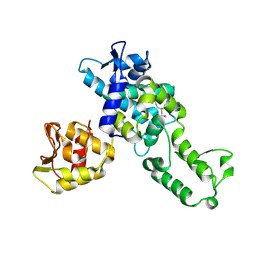 | | NEDD4 HECT with covalently bound indole-based inhibitor | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, methyl (2E)-4-{[(5-methoxy-1,2-dimethyl-1H-indol-3-yl)carbonyl]amino}but-2-enoate | | Authors: | Span, I, Smith, A.T, Kathman, S, Statsyuk, A.V, Rosenzweig, A.C. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism.
J. Am. Chem. Soc., 137, 2015
|
|
6FLP
 
 | |
1LYQ
 
 | | Crystal Structure of PcoC, a Methionine Rich Copper Resistance Protein from Escherichia coli | | Descriptor: | GLYCEROL, PcoC copper resistance protein | | Authors: | Wernimont, A.K, Huffman, D.L, Finney, L.A, Demeler, B, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2002-06-07 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and dimerization equilibria of PcoC, a methionine-rich copper resistance protein from Escherichia coli
J.BIOL.INORG.CHEM., 8, 2003
|
|
1WQ1
 
 | | RAS-RASGAP COMPLEX | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, H-RAS, ... | | Authors: | Scheffzek, K, Ahmadian, M.R, Kabsch, W, Wiesmueller, L, Lautwein, A, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants.
Science, 277, 1997
|
|
1N68
 
 | | Copper bound to the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-CL-CU LINKAGE | | Authors: | Roberts, S.A, Wildner, G.F, Grass, G, Weichsel, A, Ambrus, A, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-11-08 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Labile Regulatory Copper Ion Lies Near the T1 Copper Site in the Multicopper Oxidase CueO.
J.Biol.Chem., 278, 2003
|
|
1KV7
 
 | | Crystal Structure of CueO, a multi-copper oxidase from E. coli involved in copper homeostasis | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, PROBABLE BLUE-COPPER PROTEIN YACK | | Authors: | Roberts, S.A, Weichsel, A, Grass, G, Thakali, K, Hazzard, J.T, Tollin, G, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4M1H
 
 | | X-ray crystal structure of Chlamydia trachomatis apo NrdB | | Descriptor: | Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Basis for Assembly of the Mn(IV)/Fe(III) Cofactor in the Class Ic Ribonucleotide Reductase from Chlamydia trachomatis.
Biochemistry, 52, 2013
|
|
1KOI
 
 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH NITRIC OXIDE AT 1.08 A RESOLUTION | | Descriptor: | NITRIC OXIDE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qiu, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1X8O
 
 | | 1.01 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Nitric Oxide at pH 5.6 | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8Q
 
 | | 0.85 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus in Complex with Water at pH 5.6 | | Descriptor: | Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8P
 
 | | 0.85 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Ammonia at pH 7.4 | | Descriptor: | AMMONIA, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
1X8N
 
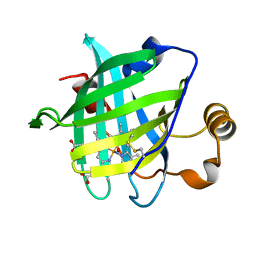 | | 1.08 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Nitric Oxide at pH 7.4 | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
4M1I
 
 | |
1U0X
 
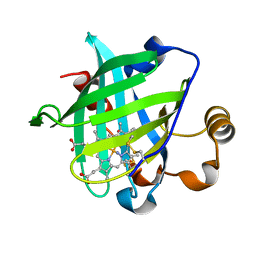 | | Crystal structure of nitrophorin 4 under pressure of xenon (200 psi) | | Descriptor: | AMMONIA, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nienhaus, K, Maes, E.M, Weichsel, A, Montfort, W.R, Nienhaus, G.U. | | Deposit date: | 2004-07-14 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural dynamics controls nitric oxide affinity in nitrophorin 4
J.Biol.Chem., 279, 2004
|
|
6FVU
 
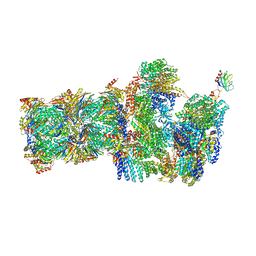 | | 26S proteasome, s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVY
 
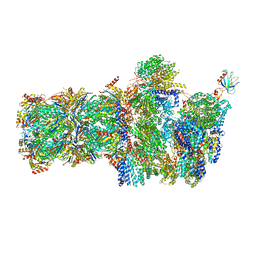 | | 26S proteasome, s6 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVW
 
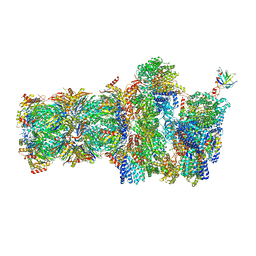 | | 26S proteasome, s4 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
