8SBS
 
 | |
6X71
 
 | |
8VWT
 
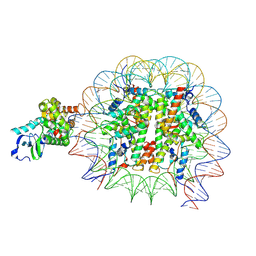 | | OGG1 bound to a nucleosome containing 8oxoG at SHL-6 (composite map) | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWU
 
 | | Nucleosome containing 8oxoG at SHL4 | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWV
 
 | | OGG1 bound to a nucleosome containing 8oxoG at SHL4 (composite map) | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWS
 
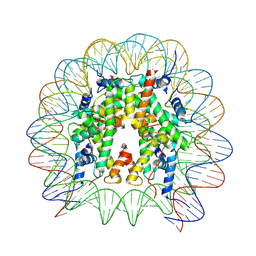 | | Nucleosome containing 8oxoG at SHL-6 | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
6PZL
 
 | |
6PYK
 
 | |
6Q0P
 
 | |
1FUR
 
 | |
6X76
 
 | | Rev1 L325G Mn2+-facilitated Product Complex with second dCTP bound | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X72
 
 | | Rev1 Mg2+-facilitated Product Complex with two monophosphates | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C*)-3'), DNA repair protein REV1, ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X75
 
 | | Rev1 Mn2+-facilitated Product Complex with second dCTP bound | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X6Z
 
 | | Rev1 Ternary Complex with dCTP and Ca2+ | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X73
 
 | | Rev1 Mg2+-facilitated Product Complex with one monophosphate | | Descriptor: | AMMONIUM ION, DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X70
 
 | | Rev1-DNA Binary Complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*G)-3'), DNA repair protein REV1, ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X77
 
 | | Rev1 R518A Ternary Complex with dCTP and Ca2+ | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X74
 
 | | Rev1 Mg2+-facilitated Product Complex with no monophosphates | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*GP*GP*GP*TP*GP*TP*GP*GP*TP*AP*GP*C*)-3'), DNA (5'-D(P*AP*TP*CP*GP*CP*TP*AP*CP*CP*AP*CP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Weaver, T.M, Freudenthal, B.D. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Visualizing Rev1 catalyze protein-template DNA synthesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9DWK
 
 | |
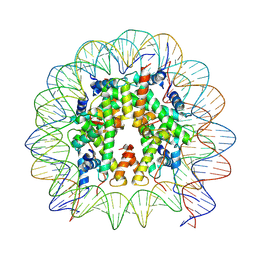
9DWF
 
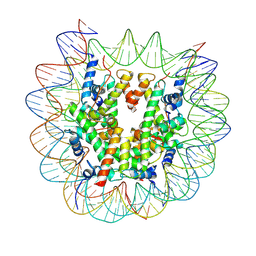 | |
9DWJ
 
 | |
9DWI
 
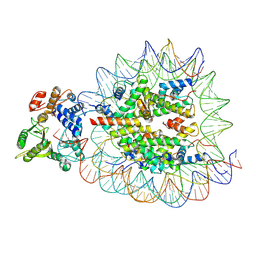 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 3, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 4, composite)
To Be Published
|
|
9DWH
 
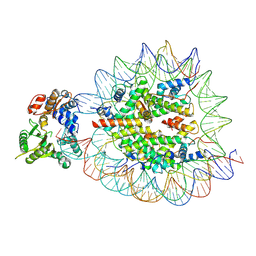 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 2, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 2, composite)
To Be Published
|
|
9DWL
 
 | |
9DWG
 
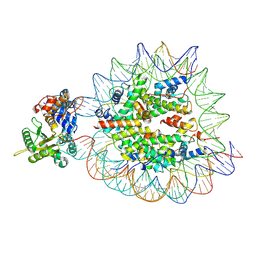 | | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 1, composite) | | Descriptor: | 601 I strand (damaged strand 1), 601 J strand (non-damaged strand), 601 K strand (damaged strand 2), ... | | Authors: | Weaver, T.M, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2024-10-09 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA Polymerase Beta bound to a nucleosome containing a 1-nt gap at SHL-4.5 (State 1, composite)
To Be Published
|
|
