3VYS
 
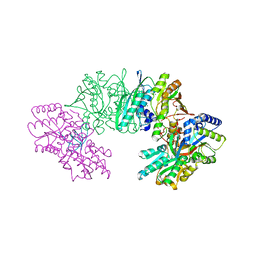 | | Crystal structure of the HypC-HypD-HypE complex (form I) | | 分子名称: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | 著者 | Watanabe, S, Miki, K. | | 登録日 | 2012-10-02 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VYU
 
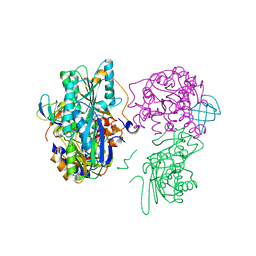 | | Crystal structure of the HypC-HypD-HypE complex (form II) | | 分子名称: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | 著者 | Watanabe, S, Miki, K. | | 登録日 | 2012-10-02 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3A43
 
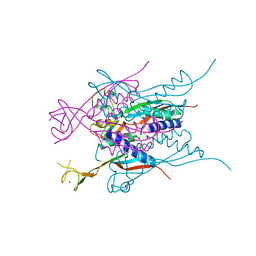 | | Crystal structure of HypA | | 分子名称: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | 著者 | Watanabe, S, Arai, T, Matsumi, R, Aromi, H, Imanaka, T, Miki, K. | | 登録日 | 2009-06-30 | | 公開日 | 2009-10-06 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
2Z1E
 
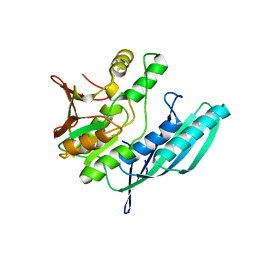 | | Crystal structure of HypE from Thermococcus kodakaraensis (outward form) | | 分子名称: | Hydrogenase expression/formation protein HypE | | 著者 | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2007-05-08 | | 公開日 | 2007-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z1C
 
 | | Crystal structure of HypC from Thermococcus kodakaraensis KOD1 | | 分子名称: | GLYCEROL, Hydrogenase expression/formation protein HypC, TETRAETHYLENE GLYCOL | | 著者 | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2007-05-08 | | 公開日 | 2007-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2ZHG
 
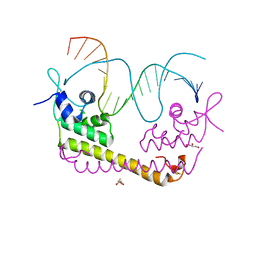 | | Crystal structure of SoxR in complex with DNA | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*DGP*DCP*DCP*DTP*DCP*DAP*DAP*DGP*DTP*DTP*DAP*DAP*DCP*DTP*DTP*DGP*DAP*DGP*DGP*DC)-3'), FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | 登録日 | 2008-02-05 | | 公開日 | 2008-03-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z1F
 
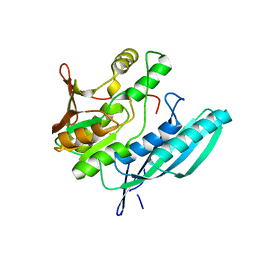 | | Crystal structure of HypE from Thermococcus kodakaraensis (inward form) | | 分子名称: | Hydrogenase expression/formation protein HypE | | 著者 | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2007-05-08 | | 公開日 | 2007-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
2Z1D
 
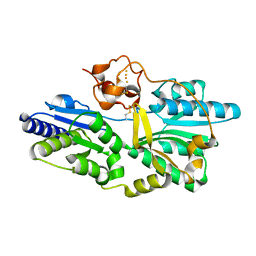 | | Crystal structure of [NiFe] hydrogenase maturation protein, HypD from Thermococcus kodakaraensis | | 分子名称: | Hydrogenase expression/formation protein hypD, IRON/SULFUR CLUSTER | | 著者 | Watanabe, S, Matsumi, R, Arai, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2007-05-08 | | 公開日 | 2007-07-17 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal Structures of [NiFe] Hydrogenase Maturation Proteins HypC, HypD, and HypE: Insights into Cyanation Reaction by Thiol Redox Signaling
Mol.Cell, 27, 2007
|
|
3A44
 
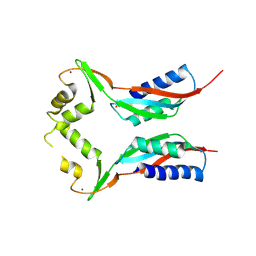 | | Crystal structure of HypA in the dimeric form | | 分子名称: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | 著者 | Watanabe, S, Arai, T, Matsumi, R, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2009-06-30 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
2ZHH
 
 | | Crystal structure of SoxR | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, FE2/S2 (INORGANIC) CLUSTER, Redox-sensitive transcriptional activator soxR | | 著者 | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | 登録日 | 2008-02-05 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8GST
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | 分子名称: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | 著者 | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | 分子名称: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | 著者 | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
2LC2
 
 | | Solution structure of the RXLR effector P. capsici AVR3a4 | | 分子名称: | AVR3a4 | | 著者 | Li, H, Koshiba, S, Yaeno, T, Sato, M, Watanabe, S, Harada, T, Shirasu, K, Kigawa, T. | | 登録日 | 2011-04-12 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A PIP-binding interface in the oomycete RXLR effector AVR3A is required for its accumulation in host cells to modulate plant immunity
To be Published
|
|
2KUQ
 
 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | 分子名称: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | 著者 | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2010-02-24 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | 分子名称: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | 著者 | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-06-24 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
6L06
 
 | |
6L07
 
 | |
7CGQ
 
 | |
7CNQ
 
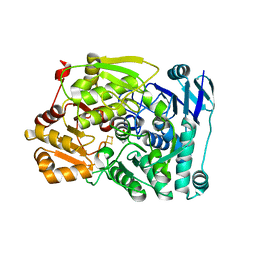 | | Crystal structure of Agrobacterium tumefaciens aconitase X (holo-form) | | 分子名称: | (2~{S},3~{R})-3-oxidanylpyrrolidine-2-carboxylic acid, FE2/S2 (INORGANIC) CLUSTER, cis-3-hydroxy-L-proline dehydratase | | 著者 | Murase, Y, Watanabe, Y, Watanabe, S. | | 登録日 | 2020-08-03 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7CNR
 
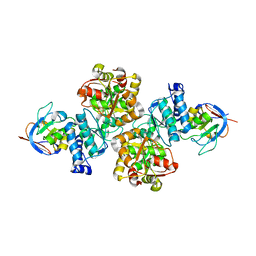 | |
7CNP
 
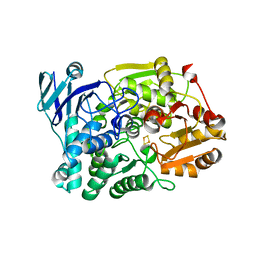 | |
7CNS
 
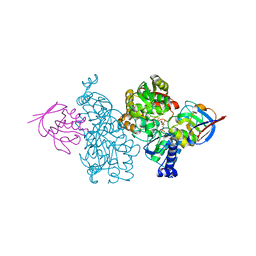 | | Crystal structure of Thermococcus kodakaraensis aconitase X (holo-form) | | 分子名称: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, DUF521 domain-containing protein, FE3-S4 CLUSTER, ... | | 著者 | Murase, Y, Watanabe, Y, Watanabe, S. | | 登録日 | 2020-08-03 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Crystal structures of aconitase X enzymes from bacteria and archaea provide insights into the molecular evolution of the aconitase superfamily.
Commun Biol, 4, 2021
|
|
7DO6
 
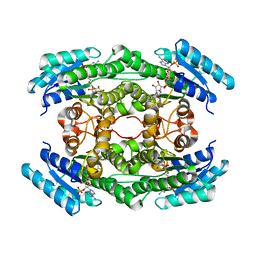 | |
8XWK
 
 | |
8Y4B
 
 | |
