3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
2Q5E
 
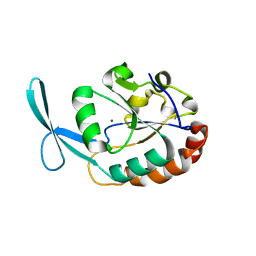 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KQ5
 
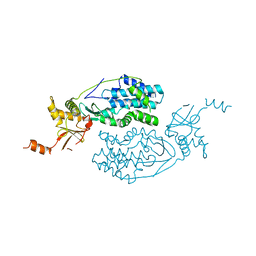 | | Crystal structure of an uncharacterized protein from Coxiella burnetii | | Descriptor: | Hypothetical cytosolic protein | | Authors: | Bonanno, J.B, Freeman, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-17 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Coxiella burnetii
To be Published
|
|
3KKE
 
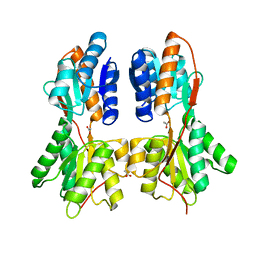 | | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, LacI family Transcriptional regulator | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Do, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis
To be Published
|
|
6C2R
 
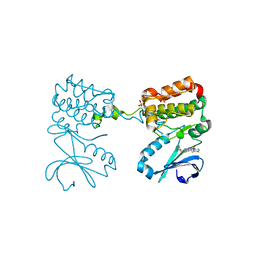 | | Aurora A ligand complex | | Descriptor: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
6C2T
 
 | | Aurora A ligand complex | | Descriptor: | (2S,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-2-methyl-4-({3-[(1,3-thiazol-2-yl)amino]isoquinolin-1-yl}methyl)piperidine-4-carboxylic acid, Aurora kinase A, DIMETHYL SULFOXIDE, ... | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
3BIL
 
 | | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum | | Descriptor: | Probable LacI-family transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum.
To be Published
|
|
2PNW
 
 | | Crystal structure of membrane-bound lytic murein transglycosylase from Agrobacterium tumefaciens | | Descriptor: | Membrane-bound lytic murein transglycosylase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of membrane-bound lytic murein transglycosylase from Agrobacterium tumefaciens.
To be Published
|
|
3C8T
 
 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
2QYA
 
 | | Crystal structure of an uncharacterized conserved protein from Methanopyrus kandleri | | Descriptor: | Uncharacterized conserved protein | | Authors: | Bonanno, J.B, Zhang, A, Bain, K.T, Adams, J, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Methanopyrus kandleri.
To be Published
|
|
3BS4
 
 | | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide | | Descriptor: | Uncharacterized protein PH0321, Unknown peptide | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide.
To be Published
|
|
3KGZ
 
 | | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris | | Descriptor: | Cupin 2 conserved barrel domain protein, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris
To be Published
|
|
3BHW
 
 | | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum.
To be Published
|
|
3BGH
 
 | | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori | | Descriptor: | Putative neuraminyllactose-binding hemagglutinin homolog, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, J, Bain, K.T, McKenzie, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori.
To be Published
|
|
3BRS
 
 | | Crystal structure of sugar transporter from Clostridium phytofermentans | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Meyers, A.J, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sugar transporter from Clostridium phytofermentans.
To be Published
|
|
3NF5
 
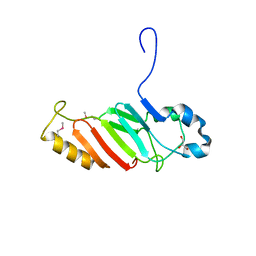 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | Descriptor: | Putative isochorismatase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
3B40
 
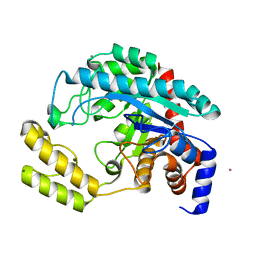 | | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa | | Descriptor: | CADMIUM ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Dickey, M, Bain, K.T, Mendoza, M, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable dipeptidase PvdM from Pseudomonas aeruginosa.
To be Published
|
|
3B5M
 
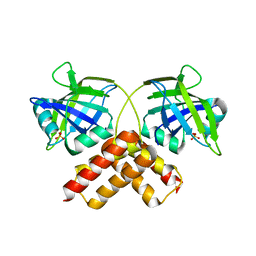 | | Crystal structure of conserved uncharacterized protein from Rhodopirellula baltica | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Bonanno, J.B, Sridhar, V, Rutter, M, Powell, A, Maletic, M, Rodgers, R, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Conserved Uncharacterized Protein from Rhodopirellula baltica.
To be Published
|
|
3EEZ
 
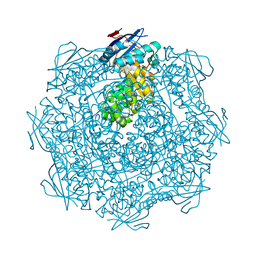 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Silicibacter pomeroyi | | Descriptor: | putative Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-07 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Silicibacter pomeroyi
To be Published
|
|
3EMU
 
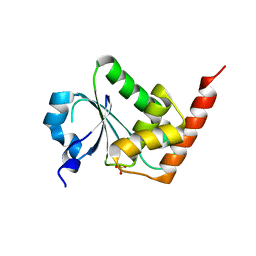 | | Crystal structure of a leucine rich repeat and phosphatase domain containing protein from Entamoeba histolytica | | Descriptor: | SULFATE ION, leucine rich repeat and phosphatase domain containing protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a leucine rich repeat and phosphatase domain containing protein from Entamoeba histolytica
To be Published
|
|
3L8K
 
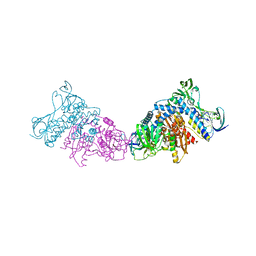 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
3ER6
 
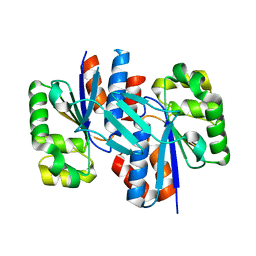 | | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus | | Descriptor: | Putative transcriptional regulator protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative transcriptional regulator protein from Vibrio parahaemolyticus
To be Published
|
|
