7EEI
 
 | |
4JE0
 
 | |
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
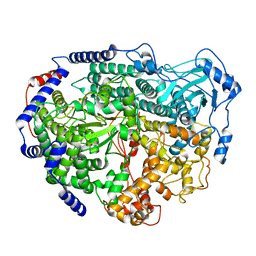
4EGT
 
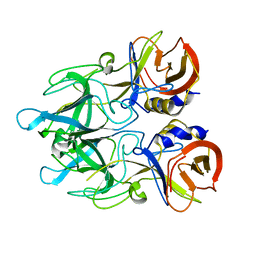 | | Crystal structure of major capsid protein P domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Xu, F, Zhang, K, Zhai, Y, Sun, F. | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
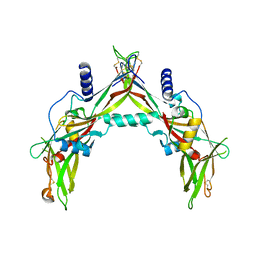
5HLZ
 
 | |
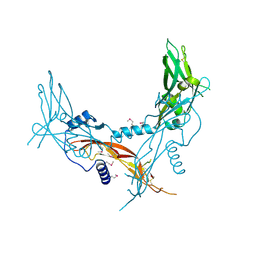
5HLY
 
 | |
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7FCE
 
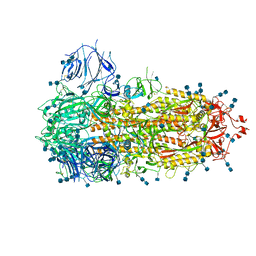 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (closed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
7FCD
 
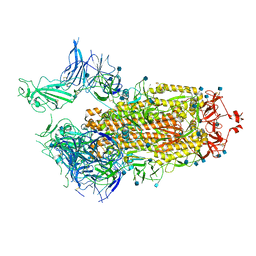 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
8WIL
 
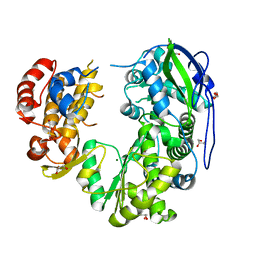 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D55 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
8WIM
 
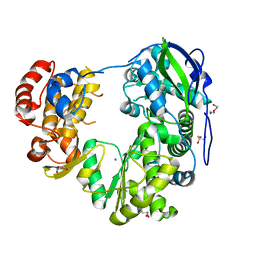 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D307 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
2L9H
 
 | | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data | | Descriptor: | C-C motif chemokine 5 | | Authors: | Wang, X, Watson, C.M, Sharp, J.S, Handel, T.M, Prestegard, J.H. | | Deposit date: | 2011-02-09 | | Release date: | 2011-06-22 | | Last modified: | 2011-08-24 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data.
Structure, 19, 2011
|
|
8VOH
 
 | |
8VOI
 
 | |
8IDO
 
 | | Crystal structure of nanobody VHH-T148 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T148, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IEE
 
 | |
8IFN
 
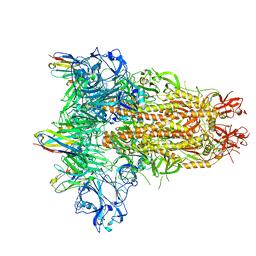 | | MERS-CoV spike trimer in complex with nanobody VHH-T148 | | Descriptor: | Spike glycoprotein, VHH-T148, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IDM
 
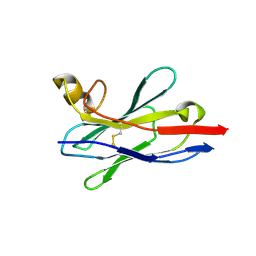 | | Crystal structure of nanobody VHH-227 with nanobody VHH-T71 and MERS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8IDI
 
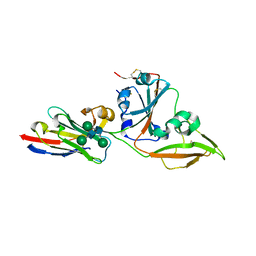 | | Crystal structure of nanobody VHH-T71 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T71, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
7CWU
 
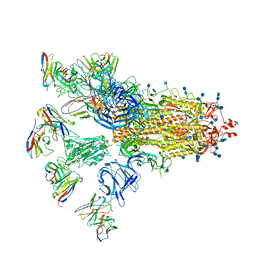 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
8WQ0
 
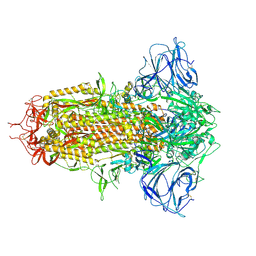 | | Cryo-EM structure of WIV1 spike glycoprotein (the closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.
J.Virol., 98, 2024
|
|
8WLZ
 
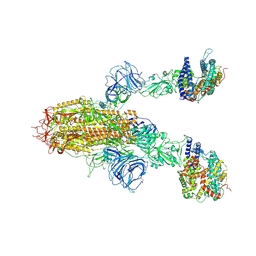 | | Cryo-EM structure of the WIV1 S-hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.
J.Virol., 98, 2024
|
|
8WLU
 
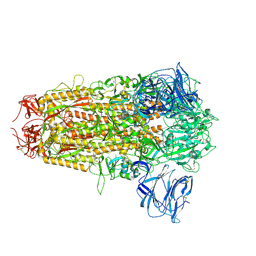 | | Cryo-EM structure of bat RsSHC014 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.
J.Virol., 98, 2024
|
|
8WLY
 
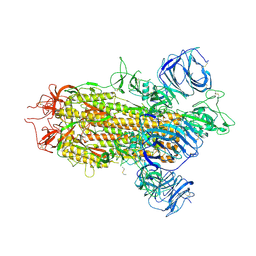 | | Cryo-EM structure of bat WIV1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.
J.Virol., 98, 2024
|
|
6LGY
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing state complexed with glycine and sodium | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, GLYCINE, NONAETHYLENE GLYCOL, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
