8SH9
 
 | | CCT G beta 5 complex best PhLP1 class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-13 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHF
 
 | | CCT-G beta 5 complex closed state 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHT
 
 | | CCT G beta 5 complex closed state 14 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHN
 
 | | CCT-G beta 5 complex closed state 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHQ
 
 | | CCT G beta 5 complex closed state 12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8SHG
 
 | | CCT G beta 5 complex closed state 9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8TOX
 
 | |
2XNB
 
 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 3,4-DIMETHYL-5-(2-{[(1Z)-4-PIPERAZIN-1-YLCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}PYRIMIDIN-4-YL)-1,3-THIAZOL-2(3H)-ONE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-08-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2XMY
 
 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 4-[4-(3,4-DIMETHYL-2-OXO-2,3-DIHYDRO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-N-(2-METHOXY-ETHYL)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
2A84
 
 | | Crystal structure of A Pantothenate synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A88
 
 | | Crystal structure of A Pantothenate synthetase, apo enzyme in C2 space group | | Descriptor: | GLYCEROL, Pantoate--beta-alanine ligase, SULFATE ION | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A7X
 
 | | Crystal Structure of A Pantothenate synthetase complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Pantoate-beta-alanine ligase, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
2A86
 
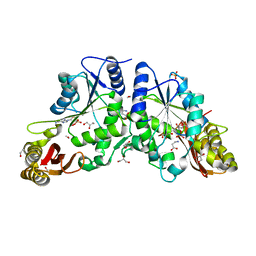 | | Crystal structure of A Pantothenate synthetase complexed with AMP and beta-alanine | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-ALANINE, ETHANOL, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-07 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pantothenate Synthetase from Mycobacterium tuberculosis, Snapshots of the Enzyme in Action.
Biochemistry, 45, 2006
|
|
1A8Y
 
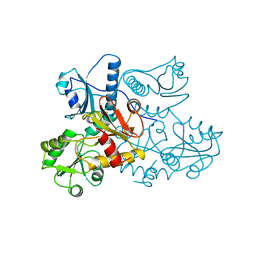 | | CRYSTAL STRUCTURE OF CALSEQUESTRIN FROM RABBIT SKELETAL MUSCLE SARCOPLASMIC RETICULUM AT 2.4 A RESOLUTION | | Descriptor: | CALSEQUESTRIN | | Authors: | Wang, S, Trumble, W.R, Liao, H, Wesson, C.R, Dunker, A.K, Kang, C. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum.
Nat.Struct.Biol., 5, 1998
|
|
7DBP
 
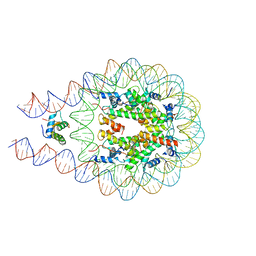 | | Linker histone defines structure and self-association behaviour of the 177 bp human chromosome | | Descriptor: | DNA (175-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Wang, S, Vogirala, K.V, Soman, A, Liu, Z.B. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Sci Rep, 11, 2021
|
|
8JRJ
 
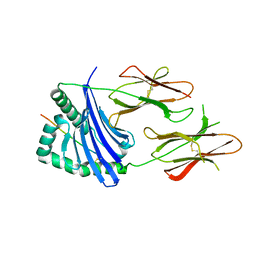 | | Crystal structure of the bat MHC II molecule at 2.8 A resolution | | Descriptor: | ALA-SER-PHE-ILE-ILE-ARG-SER-MET-PRO-GLN-GLU-THR, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, S. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the bat MHC II molecule at 2.8 A resolution
To Be Published
|
|
8JRK
 
 | | Crystal structure of the bat MHC II molecule at 2.3 A resolution | | Descriptor: | ASN-ASP-ILE-LEU-SER-ARG-LEU-ASP-PRO-PRO-GLU-ALA-SER, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, S. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bat MHC II molecule at 2.3 A resolution
To Be Published
|
|
8JR4
 
 | | Molecular structure of bat MHC II at 2.4 A resolution | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, MHC class II histocompatibility antigen, ... | | Authors: | Wang, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of bat MHC II at 2.4 A resolution
To Be Published
|
|
8JV0
 
 | |
8K8I
 
 | | De novo design protein -N14 | | Descriptor: | De novo design protein -N14 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design protein -N14
To Be Published
|
|
8K8F
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo design protein -N7
To Be Published
|
|
8K7Z
 
 | | De novo design protein -N1 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | De novo design protein -N1
To Be Published
|
|
8K7M
 
 | | De novo design protein -T01 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo design protein -T01
To Be Published
|
|
8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design protein -T03
To Be Published
|
|
8K83
 
 | | De novo design protein -N2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, De novo design protein, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design protein -N2
To Be Published
|
|
