5ZCJ
 
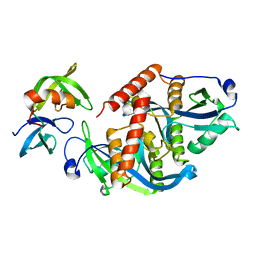 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
3CXQ
 
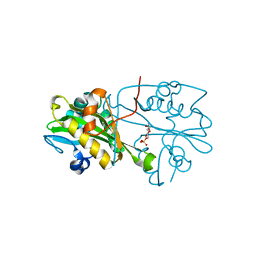 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
5ZOV
 
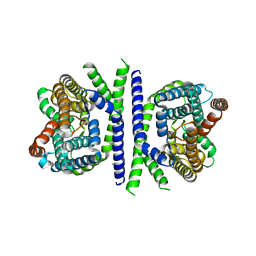 | | Inward-facing conformation of L-ascorbate transporter UlaA | | Descriptor: | ASCORBIC ACID, CALCIUM ION, PTS ascorbate-specific subunit IIBC | | Authors: | Wang, J.W. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.333 Å) | | Cite: | Inward-facing conformation of l-ascorbate transporter suggests an elevator mechanism
Cell Discov, 4, 2018
|
|
6LP3
 
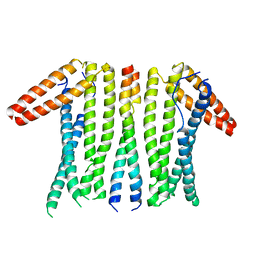 | | Structural basis and functional analysis epo1-bem3p complex for bud growth | | Descriptor: | GTPase-activating protein BEM3, Uncharacterized protein YMR124W | | Authors: | Wang, J, Li, L, Ming, Z.H, Wu, L.J, Yan, L.M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Structural basis and functional analysis epo1-bem3p complex for bud growth
To Be Published
|
|
7DKZ
 
 | | Structure of plant photosystem I-light harvesting complex I supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, J, Yu, L.J, Wang, W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structure of plant photosystem I-light harvesting complex I supercomplex at 2.4 angstrom resolution.
J Integr Plant Biol, 63, 2021
|
|
7XNO
 
 | |
5TMC
 
 | | Re-refinement of Thermus thermopiles DNA-directed RNA polymerase structure | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, J. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-23 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | On the validation of crystallographic symmetry and the quality of structures.
Protein Sci., 24, 2015
|
|
3PLS
 
 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
6BIE
 
 | |
6CWE
 
 | | Structure of alpha-GSA[8,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(6-phenylhexyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycolipid recognition by iNKT cells
To Be Published
|
|
7VQ5
 
 | |
7VQ3
 
 | |
7VQ4
 
 | |
7VQ7
 
 | | The Al-bound AtALMT1 structure at pH 5 (ALMT1Al/pH5) | | Descriptor: | ALUMINUM ION, Aluminum-activated malate transporter 1 | | Authors: | Wang, J.Q. | | Deposit date: | 2021-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
2H57
 
 | | Crystal structure of human ADP-ribosylation factor-like 6 | | Descriptor: | ADP-ribosylation factor-like protein 6, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Lew, J, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human ADP-ribosylation factor-like 6 (CASP Target)
TO BE PUBLISHED
|
|
7CPY
 
 | | Lovastatin nonaketide synthase with LovC | | Descriptor: | Lovastatin nonaketide synthase, enoyl reductase component lovC, polyketide synthase component, ... | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
6LP4
 
 | |
8HQG
 
 | |
8HVM
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF07321332
To Be Published
|
|
8HVY
 
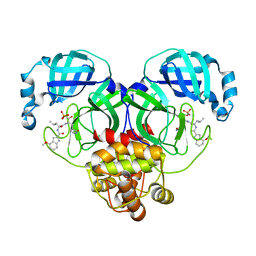 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
5YNT
 
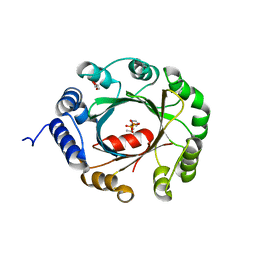 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
2KJ1
 
 | |
5YNU
 
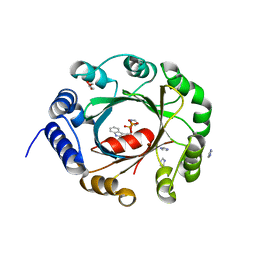 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
2KIX
 
 | |
7CPX
 
 | | Lovastatin nonaketide synthase | | Descriptor: | Lovastatin nonaketide synthase, polyketide synthase component, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
