7EJZ
 
 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7JTI
 
 | | Interphotoreceptor retinoid-binding protein (IRBP) in complex with a monoclonal antibody (F3F5 mAb5) | | Descriptor: | Retinol-binding protein 3, mAb5 Fab heavy chain, mAb5 Fab light chain | | Authors: | Sears, A.E, Albiez, S, Gulati, S, Wang, B, Kiser, P, Kovacik, L, Engel, A, Stahlberg, H, Palczewski, K. | | Deposit date: | 2020-08-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody.
Faseb J., 34, 2020
|
|
7F6Z
 
 | | Complex Structure of antibody BD-503 and RBD-501Y.V2 of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Y
 
 | | Complex Structure of antibody BD-503 and RBD-E484K of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
6J4F
 
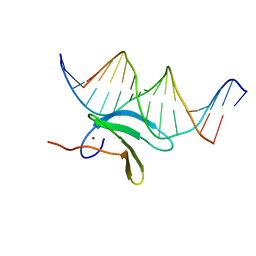 | | Crystal structure of the AtWRKY2 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 2, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the AtWRKY2 domain
To Be Published
|
|
6J4G
 
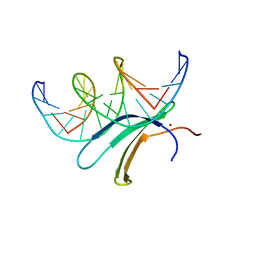 | | Crystal structure of the AtWRKY33 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 33, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the N-terminal DNA binding domain of AtWRKY33
To Be Published
|
|
6J4E
 
 | | Crystal structure of the AtWRKY1 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), WRKY transcription factor 1, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.126 Å) | | Cite: | Crystal structure of the AtWRKY1 domain
To Be Published
|
|
7VP1
 
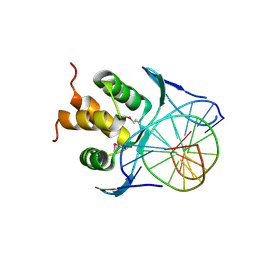 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP7
 
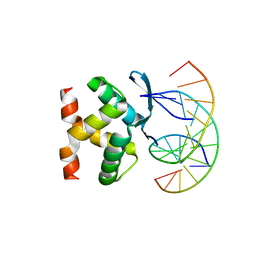 | |
7VP2
 
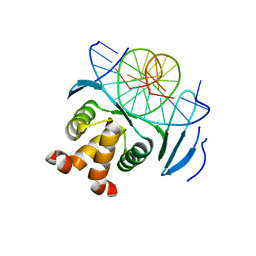 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP4
 
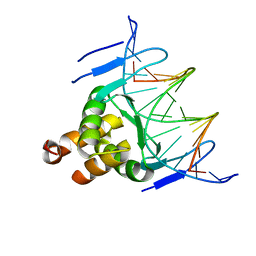 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP6
 
 | |
7W07
 
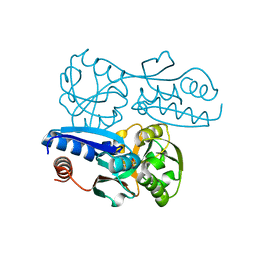 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W08
 
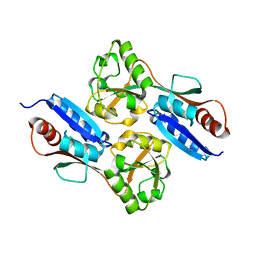 | |
7VDO
 
 | | Crystal structure of KRED F147L/L153Q/Y190P variant | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85571563 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7VE7
 
 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.720007 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7VP5
 
 | |
7VP3
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*CP*CP*AP*C)-3'), Transcription factor TCP15 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7X2R
 
 | |
7XFF
 
 | | Polysaccharide export protein Wza | | Descriptor: | Polysaccharide export protein | | Authors: | Zheng, J, Wang, B. | | Deposit date: | 2022-04-01 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polysaccharide export protein Wza
To Be Published
|
|
7XFD
 
 | | Polysaccharide export protein Wza | | Descriptor: | Polysaccharide export protein | | Authors: | Zheng, J, Wang, B. | | Deposit date: | 2022-04-01 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | polysaccharide export protein Wza
To Be Published
|
|
6M2M
 
 | |
2KSO
 
 | | EphA2:SHIP2 SAM:SAM complex | | Descriptor: | Ephrin type-A receptor 2, Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Hota, P.K, Preeti, C, Stetzik, L, Kim, S, Wang, B, Lee, H, Buck, M. | | Deposit date: | 2010-01-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the EphA2:SHIP2 SAM domain heterodimer
To be Published
|
|
7E7M
 
 | |
7EJH
 
 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F and 2-hydroxyisoindoline-1,3-dione complex | | Descriptor: | 2-oxidanylisoindole-1,3-dione, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72883928 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
