1WL0
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R44A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL3
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R91A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WKZ
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima K41A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
3VJZ
 
 | | Crystal structure of the DNA mimic protein DMP19 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wang, H.-C, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
3VK0
 
 | | Crystal Structure of hypothetical transcription factor NHTF from Neisseria | | Descriptor: | Transcriptional regulator | | Authors: | Wang, H.-C, Ko, T.-P, Wu, M.-L, Wu, H.-J, Ku, S.-C, Wang, A.H.-J. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
3EK5
 
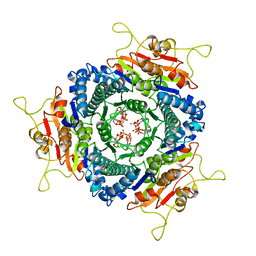 | | Unique GTP-binding Pocket and Allostery of UMP Kinase from a Gram-Negative Phytopathogen Bacterium | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Uridylate kinase | | Authors: | Tu, J.-L, Chin, K.-H, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Unique GTP-Binding Pocket and Allostery of Uridylate Kinase from a Gram-Negative Phytopathogenic Bacterium
J.Mol.Biol., 385, 2009
|
|
3EK6
 
 | |
3RO1
 
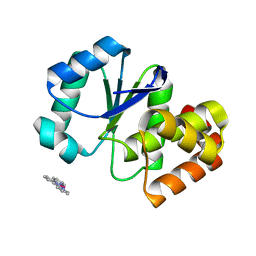 | |
3RO0
 
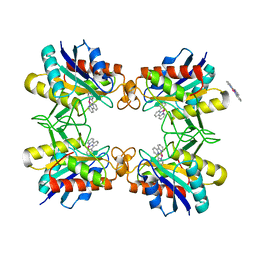 | |
3DR2
 
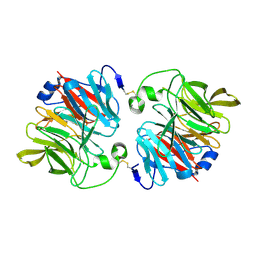 | | Structural and Functional Analyses of XC5397 from Xanthomonas campestris: A Gluconolactonase Important in Glucose Secondary Metabolic Pathways | | Descriptor: | CALCIUM ION, Exported gluconolactonase | | Authors: | Chen, C.-N, Chin, K.-H, Wang, A.H.-J, Chou, S.H. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The First Crystal Structure of Gluconolactonase Important in the Glucose Secondary Metabolic Pathways
J.Mol.Biol., 384, 2008
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1GVP
 
 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
3RNZ
 
 | |
3FWN
 
 | | Dimeric 6-phosphogluconate dehydrogenase complexed with 6-phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | Chen, Y.-Y, Ko, T.-P, Lo, L.-P, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2009-01-19 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism
J.Struct.Biol., 169, 2010
|
|
1UEH
 
 | | E. coli undecaprenyl pyrophosphate synthase in complex with Triton X-100, magnesium and sulfate | | Descriptor: | MAGNESIUM ION, OXTOXYNOL-10, SULFATE ION, ... | | Authors: | Chang, S.-Y, Ko, T.-P, Liang, P.-H, Wang, A.H.-J. | | Deposit date: | 2003-05-15 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton
J.Biol.Chem., 278, 2003
|
|
1FDG
 
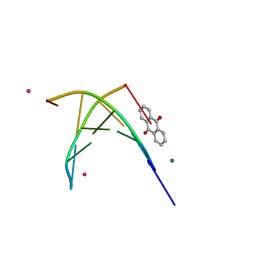 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (AMETANTRONE COMPLEX) | | Descriptor: | 1,4-BIS-[2-(2-HYDROXY-ETHYLAMINO)-ETHYLAMINO]-ANTHRAQUINONE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
1FD5
 
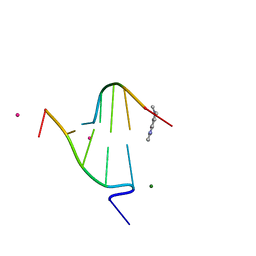 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (BISACRIDINE COMPLEX) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MACROCYCLIC-BIS-9-AMINO-ACRIDINE, ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
1FY2
 
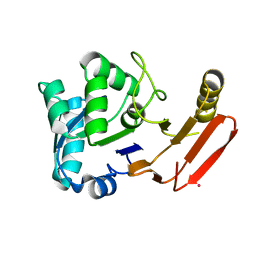 | | Aspartyl Dipeptidase | | Descriptor: | ASPARTYL DIPEPTIDASE, CADMIUM ION | | Authors: | Hakansson, K, Wang, A.H.-J, Miller, C.G. | | Deposit date: | 2000-09-28 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FYE
 
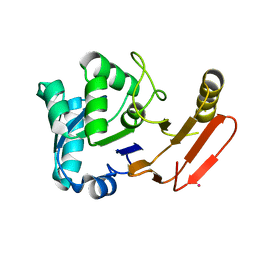 | |
1UAQ
 
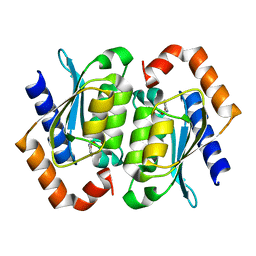 | | The crystal structure of yeast cytosine deaminase | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, ZINC ION, cytosine deaminase | | Authors: | Ko, T.-P, Lin, J.-J, Hu, C.-Y, Hsu, Y.-H, Wang, A.H.-J, Liaw, S.-H. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution
J.Biol.Chem., 278, 2003
|
|
1V7U
 
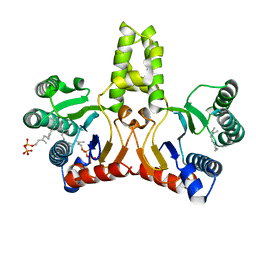 | | Crystal structure of Undecaprenyl Pyrophosphate Synthase with farnesyl pyrophosphate | | Descriptor: | FARNESYL DIPHOSPHATE, Undecaprenyl pyrophosphate synthetase | | Authors: | Chang, S.-Y, Ko, T.-P, Chen, A.P.-C, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2003-12-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies
Protein Sci., 13, 2004
|
|
1VG6
 
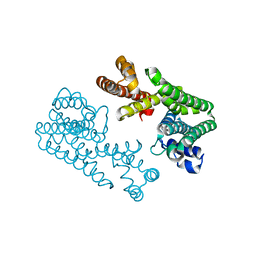 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A/I123A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG2
 
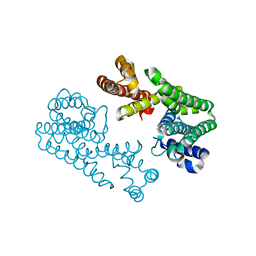 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG4
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
3LCW
 
 | | L-KDO aldolase complexed with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
