3KQX
 
 | | Structure of a protease 1 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, NONAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KR4
 
 | | Structure of a protease 3 | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4F87
 
 | | X-ray Crystal Structure of PlyCB | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8SLO
 
 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
1VCZ
 
 | |
6N87
 
 | | Plasmodium falciparum FVO apical membrane antigen 1 (AMA1) bound to MTSL spin-labelled cyclised RON2 peptide | | Descriptor: | Apical membrane antigen-1, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, backbone-cyclised peptide bcRON2hp | | Authors: | McGowan, S, Drinkwater, N. | | Deposit date: | 2018-11-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Identification of the Binding Site of Apical Membrane Antigen 1 (AMA1) Inhibitors Using a Paramagnetic Probe.
ChemMedChem, 14, 2019
|
|
1VD1
 
 | |
5WOR
 
 | |
4F88
 
 | | X-ray Crystal Structure of PlyC | | Descriptor: | PlyCA, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1VD3
 
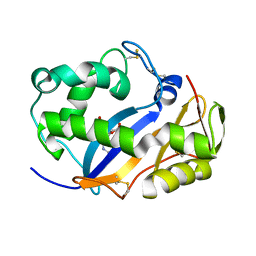 | | Ribonuclease NT in complex with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, RNase NGR3 | | Authors: | Kawano, S, Kakuta, Y, Kimura, M. | | Deposit date: | 2004-03-18 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Nicotiana glutinosa Ribonuclease NT in Complex with Nucleotide Monophosphates
to be published
|
|
6H1D
 
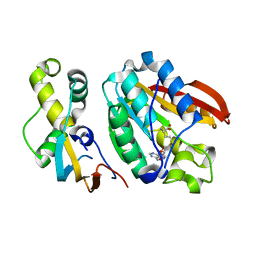 | | Crystal structure of C21orf127-TRMT112 in complex with SAH | | Descriptor: | HemK methyltransferase family member 2, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, S, Hermann, B, Metzger, E, Peng, L, Einsle, O, Schuele, R. | | Deposit date: | 2018-07-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7Y2B
 
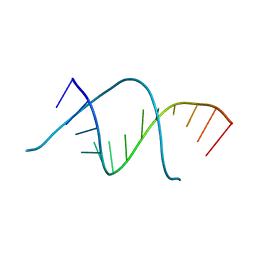 | |
3KR5
 
 | | Structure of a protease 4 | | Descriptor: | (2S)-3-[(R)-[(1S)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2OMF
 
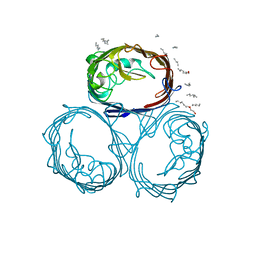 | | OMPF PORIN | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Cowan, S.W. | | Deposit date: | 1995-02-28 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Refined Structure of Ompf Porin from E.Coli at 2.4 Angstroms Resolution
To be Published
|
|
5TXD
 
 | |
5TXH
 
 | |
5H54
 
 | | Mdm12 from K. lactis 1-239 | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H55
 
 | | Mdm12 from K. lactis | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H5C
 
 | |
5DLI
 
 | | Corkscrew assembly of SOD1 residues 28-38 | | Descriptor: | GLYCEROL, IODIDE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2015-09-05 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5IIW
 
 | | Corkscrew assembly of SOD1 residues 28-38 without potassium iodide | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TXJ
 
 | |
4EME
 
 | |
4BTV
 
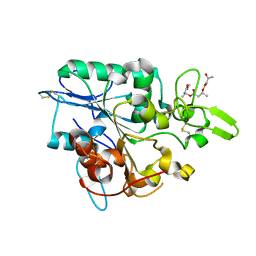 | | Structure of PhaZ7 PHB depolymerase in complex with 3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, PHB DEPOLYMERASE PHAZ7 | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4BYM
 
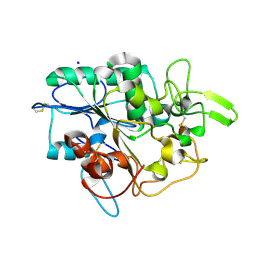 | | Structure of PhaZ7 PHB depolymerase Y105E mutant | | Descriptor: | CHLORIDE ION, PHB DEPOLYMERASE PHAZ7, SODIUM ION | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
