3VJZ
 
 | | Crystal structure of the DNA mimic protein DMP19 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wang, H.-C, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
3W1O
 
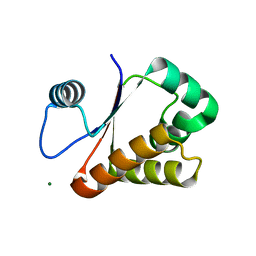 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
4XCR
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, mutant I35A | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Logan, D.T, Danielsson, J, Mu, X, Binolfi, A, Theillet, F, Bekei, B, Lang, L, Wennerstrom, H, Selenko, P, Oliveberg, M. | | Deposit date: | 2014-12-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: | Thermodynamics of protein destabilization in live cells.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
6Z1F
 
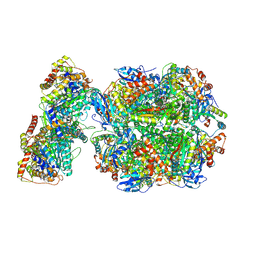 | | CryoEM structure of Rubisco Activase with its substrate Rubisco from Nostoc sp. (strain PCC7120) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, H, Bracher, A, Flecken, M, Popilka, L, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-23 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Dual Functions of a Rubisco Activase in Metabolic Repair and Recruitment to Carboxysomes.
Cell, 183, 2020
|
|
7ZSA
 
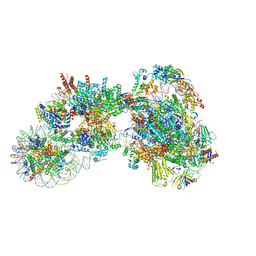 | |
7ZS9
 
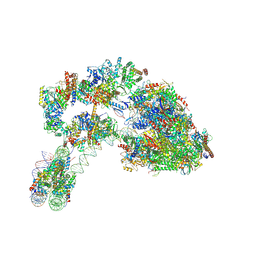 | |
3WDG
 
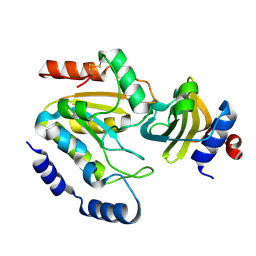 | | Staphylococcus aureus UDG / UGI complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WDF
 
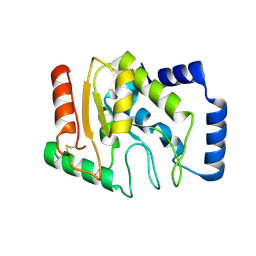 | | Staphylococcus aureus UDG | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
7MOJ
 
 | |
7MOF
 
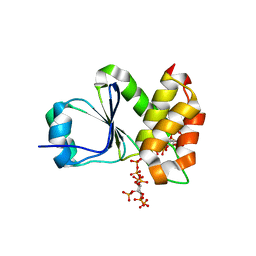 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 6-diphosphoinositol 1,2,3,4,5-pentakisphosphate 6-InsP7 | | Descriptor: | (1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, INOSITOL HEXAKISPHOSPHATE, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOE
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1)Cys150Ser in complex with 5-diphosphoinositol 1,2,3,4,6-pentakisphosphate (5-InsP7) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOG
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-PCF2 Am-InsP5, an analogue of 5-InsP7 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOI
 
 | |
7MOH
 
 | | Crystal Structure of Arabidopsis thaliana Plant and Fungi Atypical Dual Specificity Phosphatase 1(AtPFA-DSP1 ) Cys150Ser in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5PP-InsP4) and phosphate in conformation B (Pi(B)) | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, PHOSPHATE ION, Tyrosine-protein phosphatase DSP1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural expose of noncanonical molecular reactivity within the protein tyrosine phosphatase WPD loop.
Nat Commun, 13, 2022
|
|
7MOD
 
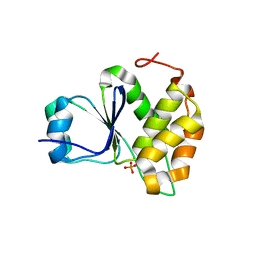 | |
7OHB
 
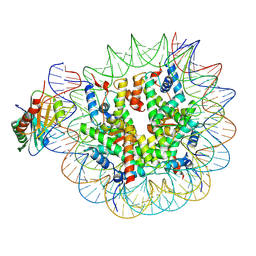 | | TBP-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHA
 
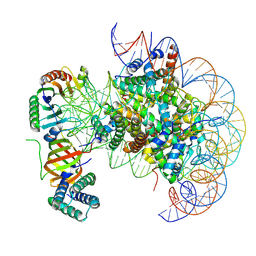 | | nucleosome with TBP and TFIIA bound at SHL +2 | | Descriptor: | DNA (122-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OH9
 
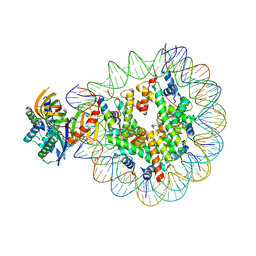 | | Nucleosome with TBP and TFIIA bound at SHL -6 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHC
 
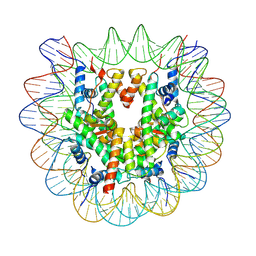 | |
7ZSB
 
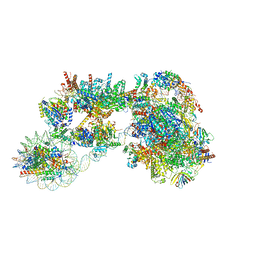 | |
5C3I
 
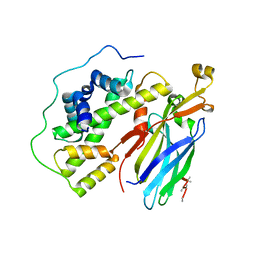 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | Descriptor: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | Authors: | Wang, H, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
2F5H
 
 | | Solution structure of the alpha-domain of human Metallothionein-3 | | Descriptor: | CADMIUM ION, Metallothionein-3 | | Authors: | Wang, H, Zhang, Q, Cai, B, Li, H.Y, Sze, K.H, Huang, Z.X, Wu, H.M, Sun, H.Z. | | Deposit date: | 2005-11-25 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of human metallothionein-3 (MT-3)
Febs Lett., 580, 2006
|
|
7OS7
 
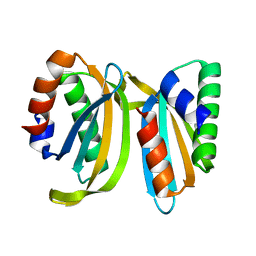 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant
To Be Published
|
|
6LMJ
 
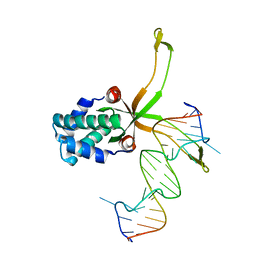 | | ASFV pA104R in complex with double-strand DNA | | Descriptor: | A104R, DNA (5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*A)-3') | | Authors: | Wang, H, Qi, J, Chai, Y, Gao, F, Liu, R. | | Deposit date: | 2019-12-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YRN
 
 | |
