3NBI
 
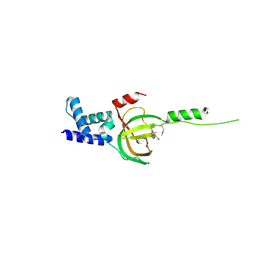 | | Crystal structure of human RMI1 N-terminus | | Descriptor: | RecQ-mediated genome instability protein 1 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3NBH
 
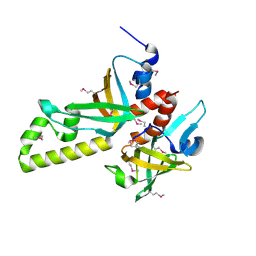 | | Crystal structure of human RMI1C-RMI2 complex | | Descriptor: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
7V9A
 
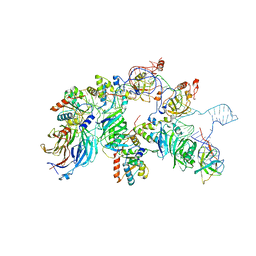 | | biogenesis module of human telomerase holoenzyme | | Descriptor: | H/ACA ribonucleoprotein complex subunit 1, H/ACA ribonucleoprotein complex subunit 2, H/ACA ribonucleoprotein complex subunit 3, ... | | Authors: | Wan, F, Ding, Y, Yang, L, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Zipper head mechanism of telomere synthesis by human telomerase.
Cell Res., 31, 2021
|
|
7V99
 
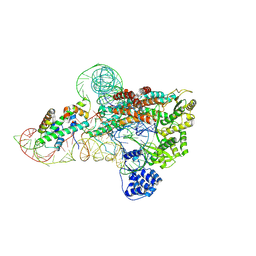 | | catalytic core of human telomerase holoenzyme | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Primer DNA, ... | | Authors: | Wan, F, Ding, Y, Yang, L, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Zipper head mechanism of telomere synthesis by human telomerase.
Cell Res., 31, 2021
|
|
6K0A
 
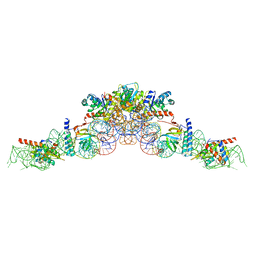 | | cryo-EM structure of an archaeal Ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
6K0B
 
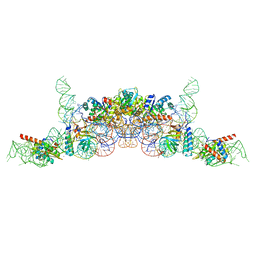 | | cryo-EM structure of archaeal Ribonuclease P with mature tRNA | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
5JJU
 
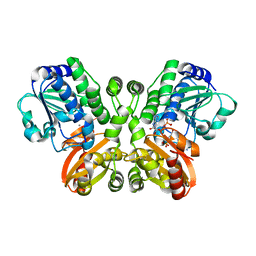 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
8D4X
 
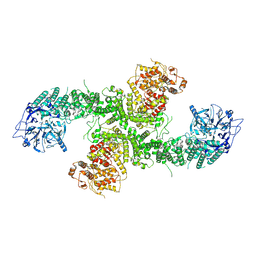 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
5WJZ
 
 | | Cryo-EM structure of B. subtilis flagellar filaments E115G | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WK5
 
 | |
5WJW
 
 | | Cryo-EM structure of B. subtilis flagellar filaments H84R | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJX
 
 | | Cryo-EM structure of B. subtilis flagellar filaments S17P | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJV
 
 | |
5WJT
 
 | |
5WK6
 
 | |
8YGZ
 
 | | The Crystal Structure of TGF beta R2 kinase domain from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TGF-beta receptor type-2 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of TGF beta R2 kinase domain from Biortus.
To Be Published
|
|
8YHP
 
 | | Structure of the PGK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Phosphoglycerate kinase 1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the PGK1 from Biortus.
To Be Published
|
|
8ZH5
 
 | |
6LIX
 
 | | CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
8YHL
 
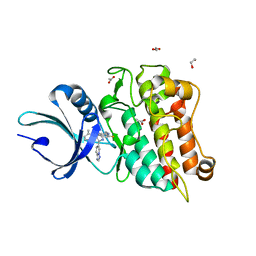 | | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, ACETATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus
To Be Published
|
|
6LIY
 
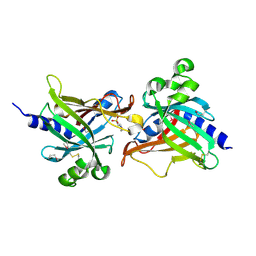 | | SeMet CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
6EF8
 
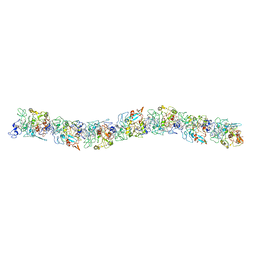 | | Cryo-EM of the OmcS nanowires from Geobacter sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C | | Authors: | Wang, F, Gu, Y, Egelman, E.H, Malvankar, N.S. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers.
Cell, 177, 2019
|
|
8ZGU
 
 | |
8YGY
 
 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
8YHH
 
 | | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus
To Be Published
|
|
