3DMD
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3DLU
 
 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | BROMIDE ION, MALONATE ION, Signal recognition particle 19 kDa protein | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3DM9
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3DM5
 
 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
6N67
 
 | |
2NG1
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-09-11 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
3DLV
 
 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | Signal recognition particle 19 kDa protein | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
1WVD
 
 | | HIV-1 Dis(Mal) Duplex CoCl2-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', COBALT (II) ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
462D
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 GENOMIC RNA DIMERIZATION INITIATION SITE | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G) -3') | | Authors: | Ennifar, E, Yusupov, M, Walter, P, Marquet, R, Ehresmann, C, Ehresmann, B, Dumas, P. | | Deposit date: | 1999-03-18 | | Release date: | 1999-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges.
Structure Fold.Des., 7, 1999
|
|
2OJ0
 
 | | Crystal structure of the duplex form of the HIV-1(LAI) RNA dimerization initiation site MN soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2007-01-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cation-dependent cleavage of the duplex form of the subtype-B HIV-1 RNA dimerization initiation site.
Nucleic Acids Res., 38, 2010
|
|
1FFH
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | FFH, MAGNESIUM ION | | Authors: | Freymann, D.M, Keenan, R.J, Stroud, R.M, Walter, P. | | Deposit date: | 1996-12-30 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the conserved GTPase domain of the signal recognition particle.
Nature, 385, 1997
|
|
1Y90
 
 | | HIV-1 Dis(Mal) Duplex Mn-Soaked | | Descriptor: | 5'-R(*CP*(5BU)P*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
1O3Z
 
 | | HIV-1 DIS(MAL) DUPLEX RU HEXAMINE-SOAKED | | Descriptor: | HIV-1 DIS(MAL) GENOMIC RNA, MAGNESIUM ION, RUTHENIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2003-05-16 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
1CQ5
 
 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | Descriptor: | SRP RNA DOMAIN IV | | Authors: | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
1RJ9
 
 | | Structure of the heterodimer of the conserved GTPase domains of the Signal Recognition Particle (Ffh) and Its Receptor (FtsY) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Signal Recognition Protein, ... | | Authors: | Egea, P.F, Shan, S.O, Napetschnig, J, Savage, D.F, Walter, P, Stroud, R.M. | | Deposit date: | 2003-11-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate twinning activates the signal recognition particle and its receptor
Nature, 427, 2004
|
|
1Y73
 
 | | HIV-1 Dis(Mal) Duplex Pt-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, PLATINUM (IV) ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
1Y95
 
 | | HIV-1 Dis(Mal) Duplex Pb-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', LEAD (II) ION, MAGNESIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
1Y6T
 
 | | HIV-1 Dis(Mal) Duplex Co Hexamine-Soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', COBALT (III) ION, SODIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
1Y99
 
 | | HIV-1 subtype A DIS RNA duplex | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Ennifar, E, Yusupov, M, Walter, P, Marquet, R, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges
Structure Fold.Des., 7, 1999
|
|
28SR
 
 | |
2B8R
 
 | | Structure oF HIV-1(LAI) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
Nat.Struct.Biol., 8, 2001
|
|
2FFH
 
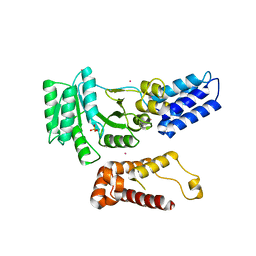 | | THE SIGNAL SEQUENCE BINDING PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | CADMIUM ION, PROTEIN (FFH), SULFATE ION | | Authors: | Keenan, R.J, Freymann, D.M, Walter, P, Stroud, R.M. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the signal sequence binding subunit of the signal recognition particle.
Cell(Cambridge,Mass.), 94, 1998
|
|
2B8S
 
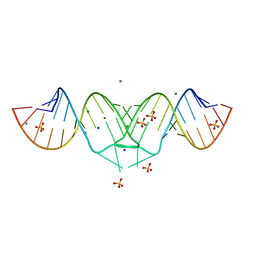 | | Structure of HIV-1(MAL) genomic RNA DIS | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Ehresmann, B, Ehresmann, C, Dumas, P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site
NAT.STRUCT.BIOL., 8, 2001
|
|
1Y6S
 
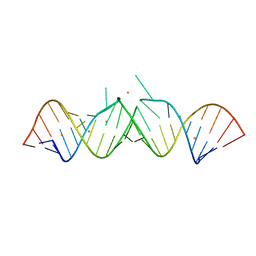 | | HIV-1 DIS(Mal) duplex Ba-soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', BARIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes
Nucleic Acids Res., 31, 2003
|
|
28SP
 
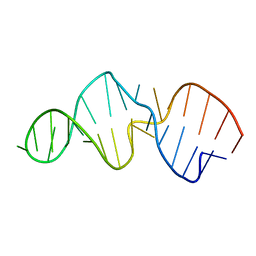 | |
