5A7H
 
 | | Comparison of the structure and activity of glycosylated and aglycosylated Human Carboxylesterase 1 | | Descriptor: | IODIDE ION, LIVER CARBOXYLESTERASE 1 | | Authors: | Arena de Souza, V, Scott, D.J, Charlton, M, Walsh, M.A, Owen, R.J. | | Deposit date: | 2015-07-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparison of the Structure and Activity of Glycosylated and Aglycosylated Human Carboxylesterase 1.
Plos One, 10, 2015
|
|
2IVG
 
 | | SITE DIRECTED MUTAGENESIS OF KEY RESIDUES INVOLVED IN THE CATALYTIC MECHANISM OF CYANASE | | Descriptor: | AZIDE ION, CHLORIDE ION, CYANATE LYASE, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, P.M. | | Deposit date: | 2006-06-13 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
8P1R
 
 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant. | | Descriptor: | 1,2-ETHANEDIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant, MAGNESIUM ION | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
8PN6
 
 | | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus | | Descriptor: | Genome polyprotein, Serine protease subunit NS2B | | Authors: | Ni, X, Fairhead, M, Balcomb, B.H, Aschenbrenner, J.C, Ferreira, L.M, Godoy, A.S, Lithgo, R.M, MacLean, E.M, Marples, P.G, Thompson, W, Tomlinson, C.W.E, Szommer, T, Wild, C, Wright, N.D, Koekemoer, L, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus
To Be Published
|
|
2IU7
 
 | |
8PXC
 
 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
2IVB
 
 | | SITE DIRECTED MUTAGENESIS OF KEY RESIDUES INVOLVED IN THE CATALYTIC MECHANISM OF CYANASE | | Descriptor: | AZIDE ION, CHLORIDE ION, CYANATE HYDRATASE, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, P.M. | | Deposit date: | 2006-06-09 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
2IVQ
 
 | |
2IV1
 
 | |
2IUO
 
 | | Site Directed Mutagenesis of Key Residues Involved in the Catalytic Mechanism of Cyanase | | Descriptor: | AZIDE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, M.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
2I9F
 
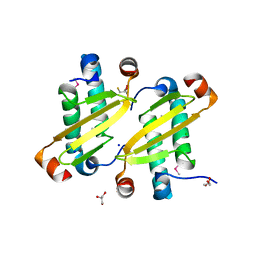 | | Structure of the equine arterivirus nucleocapsid protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Deshpande, A, Wang, S, Walsh, M, Dokland, T. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the equine arteritis virus nucleocapsid protein reveals a dimer-dimer arrangement.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4YW4
 
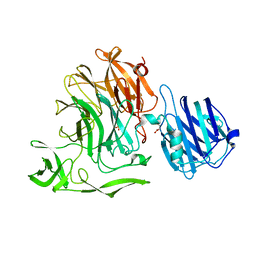 | | Streptococcus pneumoniae sialidase NanC | | Descriptor: | GLYCEROL, Neuraminidase C, PHOSPHATE ION | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel sialidase
To be published
|
|
2KWO
 
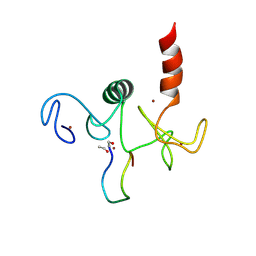 | | Solution structure of the double PHD (plant homeodomain) fingers of human transcriptional protein DPF3b bound to a histone H4 peptide containing N-terminal acetylation at Serine 1 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-14 | | Release date: | 2010-07-14 | | Last modified: | 2013-06-19 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWN
 
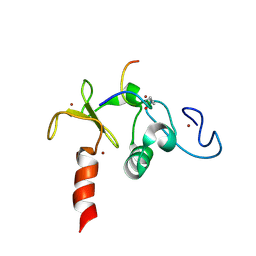 | | Solution structure of the double PHD (plant homeodomain) fingers of human transcriptional protein DPF3b bound to a histone H4 peptide containing acetylation at Lysine 16 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-13 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWK
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3b bound to a H3 peptide wild type | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWJ
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3 bound to a histone peptide containing acetylation at lysine 14 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2JKB
 
 | | Crystal structure of Streptococcus pneumoniae NanB in complex with 2, 7-anhydro-Neu5Ac | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, SIALIDASE B | | Authors: | Gut, H, King, S.J, Walsh, M.A. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Functional Studies of Streptococcus Pneumoniae Neuraminidase B: An Intramolecular Trans-Sialidase.
FEBS Lett., 582, 2008
|
|
4Y8E
 
 | | PA3825-EAL Ca-Apo Structure | | Descriptor: | CALCIUM ION, PA3825 EAL | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of PA3825 from P. aeruginosa bound to cyclic di-GMP and pGpG: new insights for a potential three-metal catalytic mechanism of EAL domains
To Be Published
|
|
7GQH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1509711879 | | Descriptor: | 6-bromo-7-hydroxy-2,2-dimethyl-2H,4H-1,3-benzodioxin-4-one, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GO5
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1198275935 | | Descriptor: | 6,8-bis(fluoranyl)chromene, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z220996120 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(1S)-1-(pyridin-3-yl)ethyl]cyclopropanecarboxamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPE
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z760031264 | | Descriptor: | 4-(1,2,4-oxadiazol-5-yl)aniline, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPK
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z905434478 | | Descriptor: | 1-[(thiophen-3-yl)methyl]piperidin-4-ol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GPX
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with NCL-00024661 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
