6VXX
 
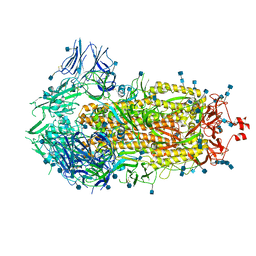 | | Structure of the SARS-CoV-2 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Wall, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Veesler, D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.
Cell, 181, 2020
|
|
6VYB
 
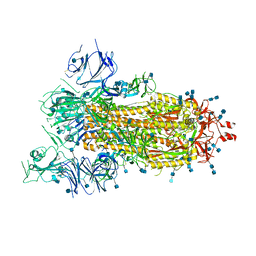 | | SARS-CoV-2 spike ectodomain structure (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Wall, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Veesler, D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.
Cell, 181, 2020
|
|
5SZS
 
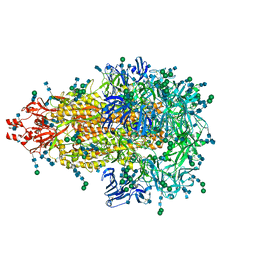 | | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Walls, A.C, Tortorici, M.A, Frenz, B, Snijder, J, Li, W, Rey, F.A, DiMaio, F, Bosch, B.J, Veesler, D. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6NB5
 
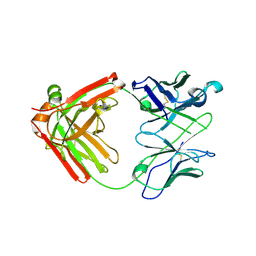 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | Descriptor: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB8
 
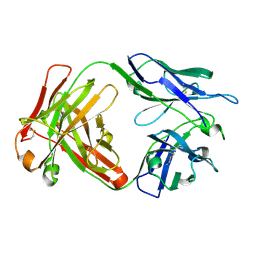 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB6
 
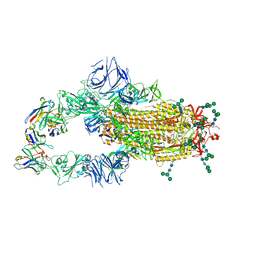 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6B3O
 
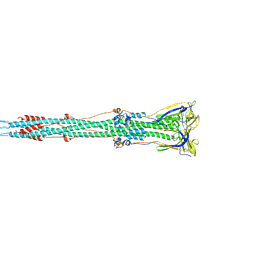 | | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Snijder, J, Xiong, X, Bosch, B.J, Rey, F.A, Veesler, D. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6NB3
 
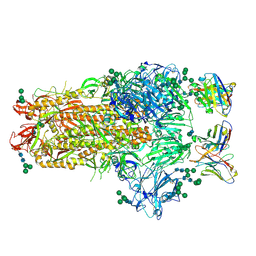 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB7
 
 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB4
 
 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
3JCL
 
 | | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer | | Descriptor: | Spike glycoprotein | | Authors: | Walls, A.C, Tortorici, M.A, Bosch, B.J, Frenz, B, Rottier, P.J.M, DiMaio, F, Rey, F.A, Veesler, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-02-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer.
Nature, 531, 2016
|
|
1QOY
 
 | | E.coli Hemolysin E (HlyE, ClyA, SheA) | | Descriptor: | HEMOLYSIN E, SULFATE ION | | Authors: | Wallace, A.J, Stillman, T.J, Atkins, A, Jamieson, S.J, Bullough, P.A, Green, J, Artymiuk, P.J. | | Deposit date: | 1999-11-25 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E. Coli Hemolysin E (Hlye, Clya, Shea): X-Ray Crystal Structure of the Toxin and Observation of Membrane Pores by Electron Microscopy
Cell(Cambridge,Mass.), 100, 2000
|
|
7JW0
 
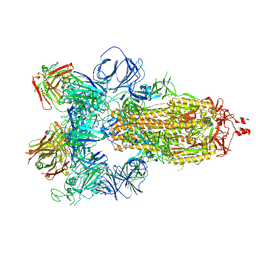 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7T8P
 
 | | CRYSTAL STRUCTURE OF T151V CAO1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase 1, FE (II) ION | | Authors: | Daruwalla, A, Kiser, P.D. | | Deposit date: | 2021-12-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF T151V CAO1
To Be Published
|
|
7T8Q
 
 | | CRYSTAL STRUCTURE OF T151G CAO1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase 1, FE (II) ION | | Authors: | Daruwalla, A, Kiser, P.D. | | Deposit date: | 2021-12-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CRYSTAL STRUCTURE OF T151G CAO1
To Be Published
|
|
2ML6
 
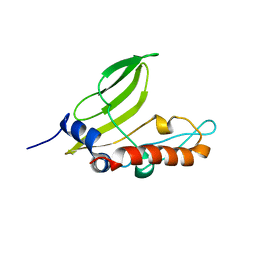 | | NMR structure of protein ZP_02069618.1 from Bacteroides uniformis ATCC 8492 | | Descriptor: | Uncharacterized protein | | Authors: | Wallmann, A, Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-02-19 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein ZP_02069618.1 from Bacteroides uniformis ATCC 8492.
To be Published
|
|
6VCG
 
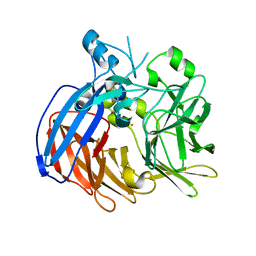 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | Descriptor: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5JPX
 
 | |
6VCH
 
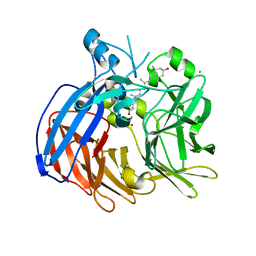 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase in complex with 3-hydroxy-beta-apo-14'-carotenal | | Descriptor: | (2E,4E,6E,8E,10E)-11-[(4R)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-5,9-dimethylundeca-2,4,6,8,10-pentaenal, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VCF
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, iron form | | Descriptor: | BICARBONATE ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7Q9B
 
 | | MHC Class I A02 Allele presenting EAAGIGILTV, in complex with Mel8 TCR | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLU-ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7Q99
 
 | | MHC Class I A02 Allele presenting NLSALGIFST, in complex with Mel5 TCR | | Descriptor: | 1,2-ETHANEDIOL, ASN-LEU-SER-ALA-LEU-GLY-ILE-PHE-SER-THR, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7Q9A
 
 | | MHC Class I A02 Allele presenting LLLGIGILVL, in complex with Mel5 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Human Mel5 T Cell Receptor, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7ZUC
 
 | | Human Major Histocompatibility Complex A2 allele presenting LLLGIGILV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-LEU-LEU-GLY-ILE-GLY-ILE-LEU-VAL, ... | | Authors: | Rizkallah, P.J, Wall, A, Sewell, A.K, Fuller, A. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7PBE
 
 | | Emergence of immune escape at dominant SARS-CoV-2 killer T-cell epitope | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, Human T-cell Receptor YLQ36, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-08-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope.
Cell, 185, 2022
|
|
