4NCS
 
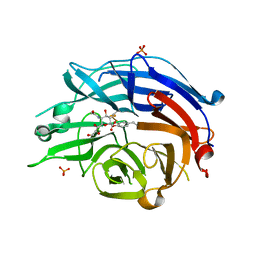 | | Human sialidase 2 in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-2,3-bis(fluoranyl)-4-oxidanyl-6-[(1S,2S)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, PHOSPHATE ION, Sialidase-2 | | Authors: | Buchini, S, Gallat, F.-X, Greig, I.R, Kim, J.-H, Wakatsuki, S, Chavas, L.M.G, Withers, S.G. | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Tuning mechanism-based inactivators of neuraminidases: mechanistic and structural insights.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2IF0
 
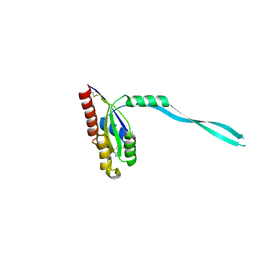 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4OWF
 
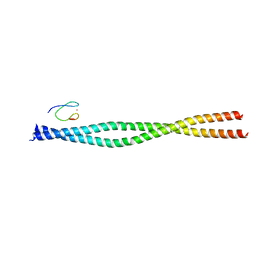 | | Crystal structure of the NEMO CoZi in complex with HOIP NZF1 domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, NF-kappa-B essential modulator, ZINC ION | | Authors: | Rahighi, S, Fujita, H, Kawasaki, M, Kato, R, Iwai, K, Wakatsuki, S. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism Underlying I kappa B Kinase Activation Mediated by the Linear Ubiquitin Chain Assembly Complex.
Mol.Cell.Biol., 34, 2014
|
|
6N6R
 
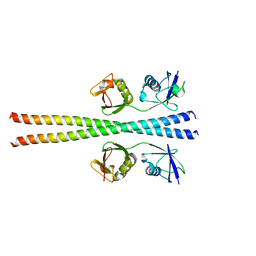 | |
6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
6N5M
 
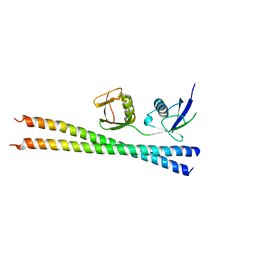 | |
6P5T
 
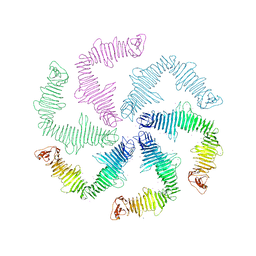 | | Surface-layer (S-layer) RsaA protein from Caulobacter crescentus bound to strontium and iodide | | Descriptor: | IODIDE ION, S-layer protein, STRONTIUM ION | | Authors: | Chan, A.C, Herrmann, J, Smit, J, Wakatsuki, S, Murphy, M.E. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bacterial surface layer protein exploits multistep crystallization for rapid self-assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Z4K
 
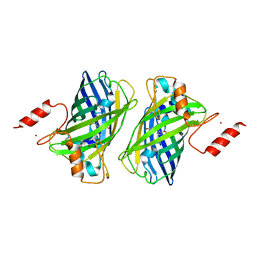 | |
4Z4M
 
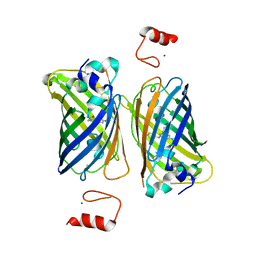 | |
3H3T
 
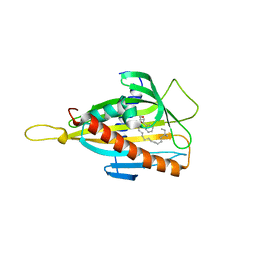 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3R
 
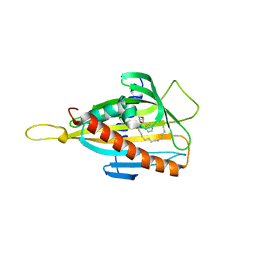 | | Crystal structure of the CERT START domain in complex with HPA-14 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tetradecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3Q
 
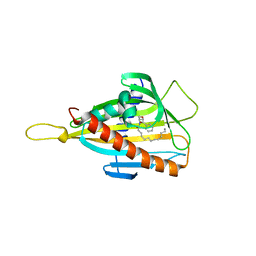 | | Crystal structure of the CERT START domain in complex with HPA-13 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tridecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3S
 
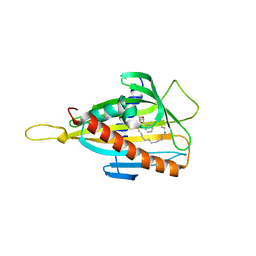 | | Crystal structure of the CERT START domain in complex with HPA-15 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]pentadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
6VA7
 
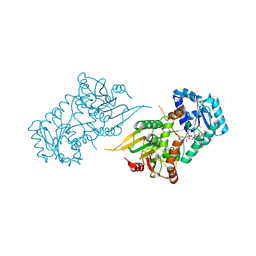 | |
6VA8
 
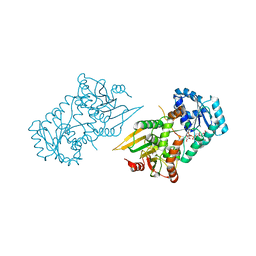 | |
6VA9
 
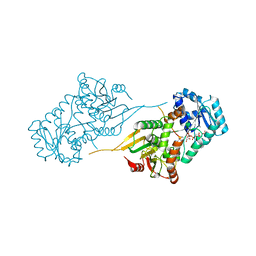 | |
6VAQ
 
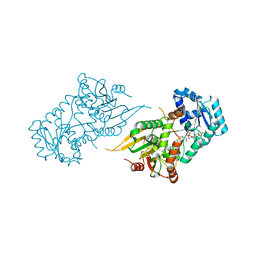 | |
6VA0
 
 | |
1EBE
 
 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
3T5W
 
 | | 2ME modified human SOD1 | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ihara, K, Yamaguchi, Y, Torigoe, H, Wakatsuki, S, Taniguchi, N, Suzuki, K, Fujiwara, N. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural switching of Cu,Zn-superoxide dismutases at loop VI: insights from the crystal structure of 2-mercaptoethanol-modified enzyme
Biosci.Rep., 32, 2012
|
|
3U3Q
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U3T
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FW9
 
 | | Crystal structure of the Lon-like protease MtaLonC | | Descriptor: | PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Ihara, K, Kuo, C.I, Huang, K.F, Wakatsuki, S. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1UJJ
 
 | | VHS domain of human GGA1 complexed with C-terminal peptide from BACE | | Descriptor: | ADP-ribosylation factor binding protein GGA1, C-terminal peptide from Beta-secretase | | Authors: | Shiba, T, Kametaka, S, Kawasaki, M, Shibata, M, Waguri, S, Uchiyama, Y, Wakatsuki, S. | | Deposit date: | 2003-08-05 | | Release date: | 2004-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Phosphoregulation of beta-Secretase Sorting Signal by the VHS Domain of GGA1
TRAFFIC, 5, 2004
|
|
1UJK
 
 | | VHS domain of human GGA1 complexed with C-terminal phosphopeptide from BACE | | Descriptor: | ADP-ribosylation factor binding protein GGA1, C-terminal peptide from Beta-secretase, IODIDE ION | | Authors: | Shiba, T, Kametaka, S, Kawasaki, M, Shibata, M, Waguri, S, Uchiyama, Y, Wakatsuki, S. | | Deposit date: | 2003-08-05 | | Release date: | 2004-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Phosphoregulation of beta-Secretase Sorting Signal by the VHS Domain of GGA1
TRAFFIC, 5, 2004
|
|
