4DIM
 
 | |
4KLK
 
 | | Phage-related protein DUF2815 from Enterococcus faecalis | | Descriptor: | ETHANOL, GLYCEROL, Phage-related protein DUF2815 | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Phage-related protein DUF2815 from Enterococcus faecalis
To be Published
|
|
1RZ3
 
 | |
3ONQ
 
 | | Crystal Structure of Regulator of Polyketide Synthase Expression BAD_0249 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, Regulator of polyketide synthase expression, SULFATE ION | | Authors: | Kim, Y, Wu, R, Tan, K, Morales, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-30 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal Structure of Regulator of Polyketide Synthase Expression BAD_0249 from Bifidobacterium adolescentis
To be Published
|
|
4XVO
 
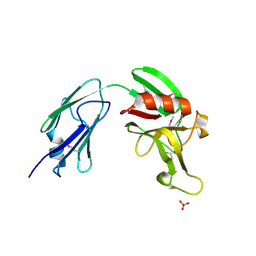 | | L,D-transpeptidase from Mycobacterium smegmatis | | Descriptor: | L,D-transpeptidase, PHOSPHATE ION | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-27 | | Release date: | 2015-02-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L,D-transpeptidase from Mycobacterium smegmatis
to be published
|
|
4ESY
 
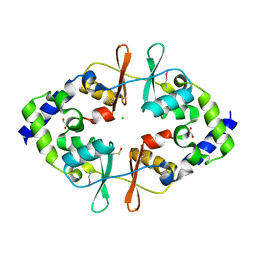 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
3LAX
 
 | |
3MUQ
 
 | |
3NJA
 
 | | The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472. | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable GGDEF family protein, ... | | Authors: | Tan, K, Wu, R, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | The crystal structure of the PAS domain of a GGDEF family protein from Chromobacterium violaceum ATCC 12472.
To be Published
|
|
5VES
 
 | | The 2.4A crystal structure of OmpA domain of OmpA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | Outer membrane protein A, SULFATE ION | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2017-04-05 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into PG-binding, conformational change, and dimerization of the OmpA C-terminal domains from Salmonella enterica serovar Typhimurium and Borrelia burgdorferi.
Protein Sci., 26, 2017
|
|
3LDU
 
 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3QSG
 
 | |
3QSJ
 
 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3R0V
 
 | | The crystal structure of an alpha/beta hydrolase from Sphaerobacter thermophilus DSM 20745. | | Descriptor: | Alpha/beta hydrolase fold protein, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | The crystal structure of an alpha/beta hydrolase from Sphaerobacter thermophilus DSM 20745.
To be Published
|
|
5UBK
 
 | | Inactive S1A/N269D-cpPvdQ mutant in complex with the pyoverdine precursor PVDIq reveals a specific binding pocket for the D-Tyr of this substrate | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, N-[(1R)-1-{(6S)-6-[(2-amino-2-oxoethyl)carbamoyl]-1,4,5,6-tetrahydropyrimidin-2-yl}-2-(4-hydroxyphenyl)ethyl]-N~2~-tetradecanoyl-L-glutamine | | Authors: | Mascarenhas, R, Catlin, D, Wu, R, Clevenger, K, Fast, W, Liu, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-01 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Circular Permutation Reveals a Chromophore Precursor Binding Pocket of the Siderophore Tailoring Enzyme PvdQ
To Be Published
|
|
4KTB
 
 | | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Tan, K, Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603
To be Published
|
|
4MK6
 
 | | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, Probable Dihydroxyacetone Kinase Regulator DHSK_reg | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e
To be Published
|
|
4MO9
 
 | | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula | | Descriptor: | GLYCEROL, Periplasmic binding protein, trimethylamine oxide | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula
To be Published
|
|
4MX8
 
 | |
4MNR
 
 | | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta
To be Published
|
|
5TTX
 
 | |
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-07-17 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
3NRV
 
 | |
5ERE
 
 | | Extracellular ligand binding receptor from Desulfohalobium retbaense DSM5692 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXO-4-METHYLPENTANOIC ACID, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Cuff, M, Wu, R, Endres, M, Pokkuluri, P.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-14 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel extracellular ligand receptor
To Be Published
|
|
1YB4
 
 | | Crystal Structure of the Tartronic Semialdehyde Reductase from Salmonella typhimurium LT2 | | Descriptor: | tartronic semialdehyde reductase | | Authors: | Kim, Y, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
