7K1V
 
 | |
7JP0
 
 | | Crystal structure of Mpro with inhibitor r1 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2R)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-07 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Mpro with inhibitor r1
To Be Published
|
|
7JPZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ5
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JYS
 
 | | hALK in complex with 3-(3-chlorophenyl)-5-methyl-1H-pyrazole | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)-5-methyl-1H-pyrazole, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7JQ3
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI6 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7JPY
 
 | |
7JQ4
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI7 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JYT
 
 | | hALK in complex with 3-(3-methyl-1H-pyrazol-5-yl)pyridine | | Descriptor: | 3-(3-methyl-1H-pyrazol-5-yl)pyridine, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7JQ0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI3 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JY4
 
 | | hALK in complex with ((1S,2S)-1-(2,4-difluorophenyl)-2-(2-(3-methyl-1H-pyrazol-5-yl)-4-(trifluoromethyl)phenoxy)cyclopropyl)methanamine | | Descriptor: | 1-{(1S,2S)-1-(2,4-difluorophenyl)-2-[2-(3-methyl-1H-pyrazol-5-yl)-4-(trifluoromethyl)phenoxy]cyclopropyl}methanamine, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7L4W
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 2d | | Descriptor: | (2s,4R)-2-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, Monoglyceride lipase | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4U
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1h | | Descriptor: | (5S)-5-(3-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidin-1-yl}-3-oxopropyl)pyrrolidin-2-one, CHLORIDE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J, Dougan, D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4T
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-2H-1,4-benzoxazin-3(4H)-one, ACETATE ION, ... | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L50
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 4f | | Descriptor: | (2s,4R)-2-{3-[(3-chloro-4-methylphenyl)methoxy]azetidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, ACETATE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7JYR
 
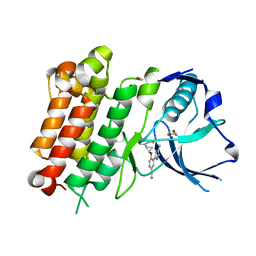 | | hALK in complex with 1-[(1R,2R)-1-(2,4-difluorophenyl)-2-[2-(5-methyl-1H-pyrazol-3-yl)-4-(trifluoromethyl)phenoxy]cyclopropyl]methanamine | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(1R,2R)-2-(2,4-difluorophenyl)cyclopropyl]oxy}-3-(5-methyl-1H-pyrazol-3-yl)benzonitrile, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7K6Q
 
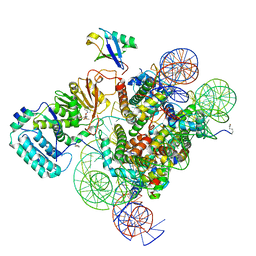 | | Active state Dot1 bound to the H4K16ac nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7K6P
 
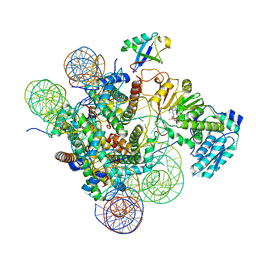 | | Active state Dot1 bound to the unacetylated H4 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
