1ITF
 
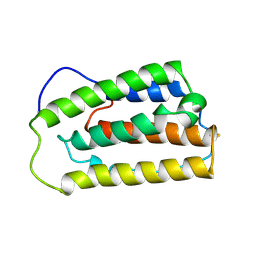 | | INTERFERON ALPHA-2A, NMR, 24 STRUCTURES | | Descriptor: | INTERFERON ALPHA-2A | | Authors: | Klaus, W, Gsell, B, Labhardt, A.M, Wipf, B, Senn, H. | | Deposit date: | 1997-08-22 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
J.Mol.Biol., 274, 1997
|
|
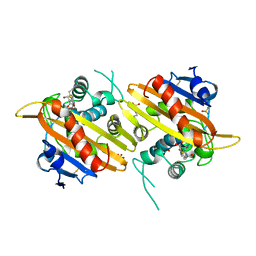
1H4U
 
 | | Domain G2 of mouse nidogen-1 | | Descriptor: | NIDOGEN-1 | | Authors: | Hopf, M, Gohring, W, Ries, A, Timpl, R, Hohenester, E. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure and Mutational Analysis of a Perlecan-Binding Fragment of Nidogen-1
Nat.Struct.Biol., 8, 2001
|
|
1H5X
 
 | |
7X4L
 
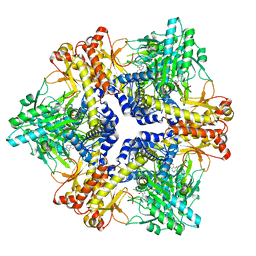 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
1INJ
 
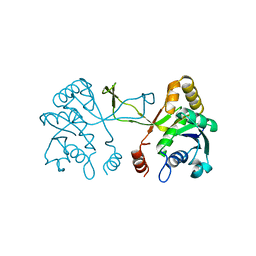 | | CRYSTAL STRUCTURE OF THE APO FORM OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, CALCIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
2DW7
 
 | | Crystal structure of D-tartrate dehydratase from Bradyrhizobium japonicum complexed with Mg++ and meso-tartrate | | Descriptor: | Bll6730 protein, MAGNESIUM ION, S,R MESO-TARTARIC ACID | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
1J8E
 
 | | Crystal structure of ligand-binding repeat CR7 from LRP | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 1 | | Authors: | Simonovic, M, Dolmer, K, Huang, W, Strickland, D.K, Volz, K, Gettins, P.G.W. | | Deposit date: | 2001-05-21 | | Release date: | 2001-12-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor.
Biochemistry, 40, 2001
|
|
1IYG
 
 | | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA | | Descriptor: | Hypothetical protein (2010003O14) | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Koshiba, S, Hamada, T, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-14 | | Release date: | 2003-02-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA
To be Published
|
|
1FYD
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE AMP, ONE PYROPHOSPHATE ION AND ONE MG2+ ION | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2000-09-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1J0N
 
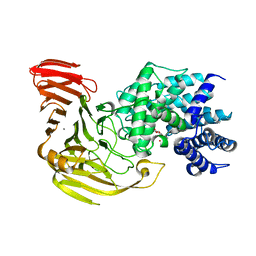 | | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan | | Descriptor: | 4,6-O-[(1S)-1-carboxyethylidene]-beta-D-glucopyranose, CALCIUM ION, XANTHAN LYASE | | Authors: | Hashimoto, W, Nankai, H, Mikami, B, Murata, K. | | Deposit date: | 2002-11-19 | | Release date: | 2003-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Bacillus sp. GL1 Xanthan Lyase, Which Acts on the Side Chains of Xanthan.
J.Biol.Chem., 278, 2003
|
|
1FU0
 
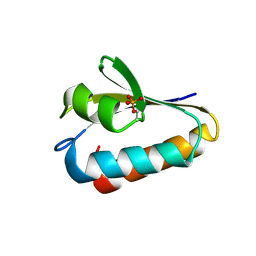 | | CRYSTAL STRUCTURE ANALYSIS OF THE PHOSPHO-SERINE 46 HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Audette, G.F, Engelmann, R, Hengstenberg, W, Deutscher, J, Hayakawa, K, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A resolution structure of phospho-serine 46 HPr from Enterococcus faecalis.
J.Mol.Biol., 303, 2000
|
|
1IHC
 
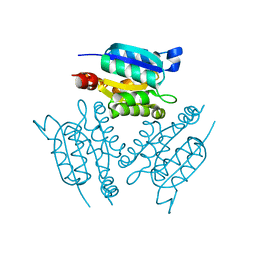 | | X-ray Structure of Gephyrin N-terminal Domain | | Descriptor: | Gephyrin | | Authors: | Sola, M, Kneussel, M, Heck, I.S, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-04-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the trimeric N-terminal domain of gephyrin.
J.Biol.Chem., 276, 2001
|
|
1J85
 
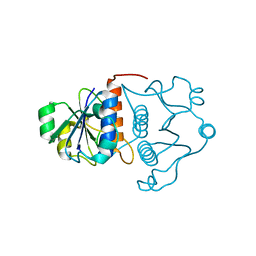 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1G27
 
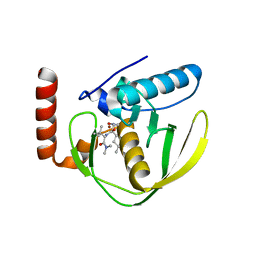 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G2P
 
 | | CRYSTAL STRUCTURE OF ADENINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | ADENINE PHOSPHORIBOSYLTRANSFERASE 1, SULFATE ION | | Authors: | Shi, W, Tanaka, K.S.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of adenine phosphoribosyltransferase from Saccharomyces cerevisiae.
Biochemistry, 40, 2001
|
|
1G4K
 
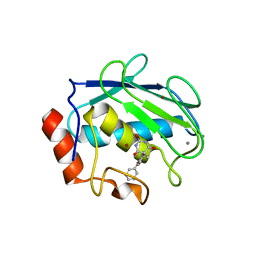 | | X-ray Structure of a Novel Matrix Metalloproteinase Inhibitor Complexed to Stromelysin | | Descriptor: | 5-METHYL-5-(4-PHENOXY-PHENYL)-PYRIMIDINE-2,4,6-TRIONE, CALCIUM ION, GLYCEROL, ... | | Authors: | Dunten, P, Kammlott, U, Crowther, R, Levin, W, Foley, L.H, Wang, P, Palermo, R. | | Deposit date: | 2000-10-27 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a novel matrix metalloproteinase inhibitor complexed to stromelysin.
Protein Sci., 10, 2001
|
|
1G8Y
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC REPLICATIVE HELICASE REPA OF PLASMID RSF1010 | | Descriptor: | REGULATORY PROTEIN REPA | | Authors: | Niedenzu, T, Roeleke, D, Bains, G, Scherzinger, E, Saenger, W. | | Deposit date: | 2000-11-21 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010.
J.Mol.Biol., 306, 2001
|
|
1JO0
 
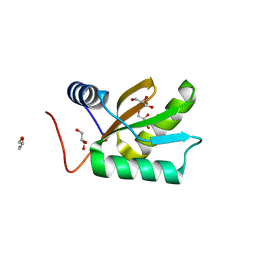 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
1FW6
 
 | | CRYSTAL STRUCTURE OF A TAQ MUTS-DNA-ADP TERNARY COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Junop, M.S, Obmolova, G, Rausch, K, Hsieh, P, Yang, W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-02-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair.
Mol.Cell, 7, 2001
|
|
1JRF
 
 | | NMR Solution Structure of the Viral Receptor Domain of Tva | | Descriptor: | CALCIUM ION, SUBGROUP A ROUS SARCOMA VIRUS RECEPTORS PG800 AND PG950 | | Authors: | Wang, Q.-Y, Huang, W, Dolmer, K, Gettins, P.G.W, Rong, L. | | Deposit date: | 2001-08-13 | | Release date: | 2002-03-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the viral receptor domain of Tva and its implications in viral entry.
J.Virol., 76, 2002
|
|
1JX4
 
 | | Crystal Structure of a Y-family DNA Polymerase in a Ternary Complex with DNA Substrates and an Incoming Nucleotide | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*TP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2001-09-05 | | Release date: | 2001-10-05 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication.
Cell(Cambridge,Mass.), 107, 2001
|
|
1HBM
 
 | | METHYL-COENZYME M REDUCTASE ENZYME PRODUCT COMPLEX | | Descriptor: | CHLORIDE ION, FACTOR 430, GLYCEROL, ... | | Authors: | Ermler, U, Grabarse, W. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
1JXZ
 
 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GYX
 
 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
