5ZL5
 
 | | Crystal structure of DFA-IIIase mutant C387A from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase C387A mutant, GLYCEROL | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKW
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with GF2 | | Descriptor: | DFA-IIIase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZLA
 
 | | Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase C387A mutant | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
1B26
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
1B3B
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-07 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
5ZKU
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | Descriptor: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKY
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lid | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKS
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
2H16
 
 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2H17
 
 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
1DAV
 
 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (20 STRUCTURES) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1DAQ
 
 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
2HIN
 
 | | Structure of N15 Cro at 1.05 A: an ortholog of lambda Cro with a completely different but equally effective dimerization mechanism | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Dubrava, M.S, Ingram, W.M, Roberts, S.A, Weichsel, A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces.
Protein Sci., 17, 2008
|
|
2HLZ
 
 | | Crystal Structure of human ketohexokinase | | Descriptor: | Ketohexokinase, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-10 | | Release date: | 2006-08-08 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Residues in the fructose-binding pocket are required for ketohexokinase-A activity.
J.Biol.Chem., 300, 2024
|
|
2I7Q
 
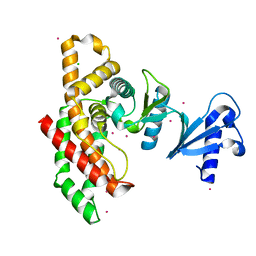 | | Crystal structure of Human Choline Kinase A | | Descriptor: | CHLORIDE ION, Choline kinase alpha, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Wasney, G, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Human Choline Kinase A
To be Published
|
|
7DL4
 
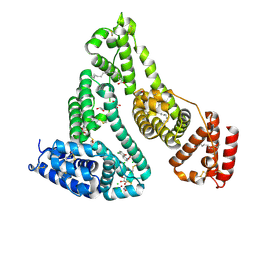 | |
6JBS
 
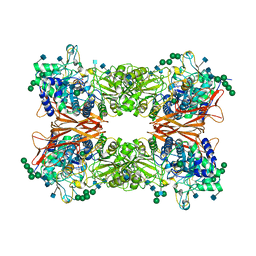 | | Bifunctional xylosidase/glucosidase LXYL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-01-26 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
6HJ6
 
 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJ5
 
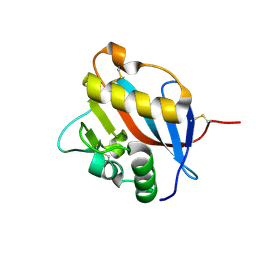 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 5.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJC
 
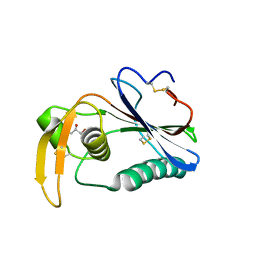 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-09-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJ4
 
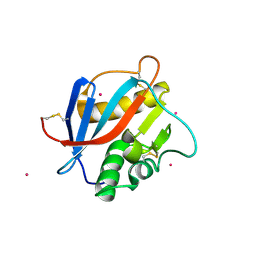 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
7EY2
 
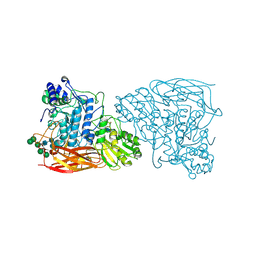 | |
7EY1
 
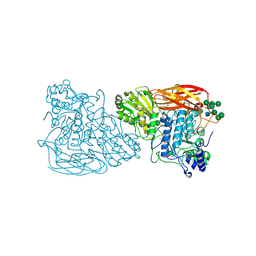 | | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Bifunctional xylosidase/glucosidase LXYL with intermediate substrate xylose
To Be Published
|
|
6KJ0
 
 | | Bifunctional xylosidase/glucosidase LXYL mutant E529Q C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, Deacetyltaxol, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-07-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
5AHE
 
 | | Crystal structure of Salmonella enterica HisA | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
