4DZY
 
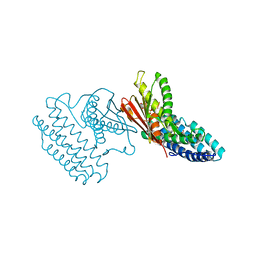 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with ADP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DFF
 
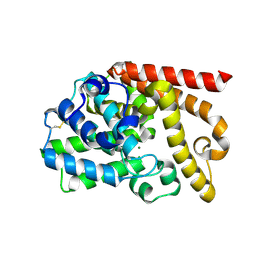 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | Descriptor: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-14 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DPX
 
 | |
1NI2
 
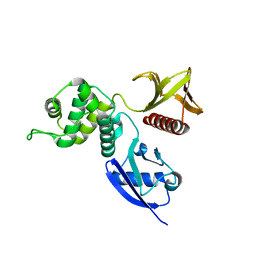 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
4AYC
 
 | | RNF8 RING domain structure | | Descriptor: | CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF8, GLYCEROL, ... | | Authors: | Mattiroli, F, Vissers, J.H.A, Van Dijk, W.J, Ikpa, P, Citterio, E, Vermeulen, W, Marteijn, J.A, Sixma, T.K. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rnf168 Ubiquitinates K13-15 on H2A/H2Ax to Drive DNA Damage Signaling
Cell(Cambridge,Mass.), 150, 2012
|
|
4CCY
 
 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
1SQA
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-4-(PYRIMIDIN-2-YLAMINO)-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4EO8
 
 | | HCV NS5B polymerase inhibitors: Tri-substituted acylhydrazines as tertiary amide bioisosteres | | Descriptor: | 5-(3,3-dimethylbut-1-yn-1-yl)-3-{2,2-dimethyl-1-[(trans-4-methylcyclohexyl)carbonyl]hydrazinyl}thiophene-2-carboxylic acid, RNA-directed RNA polymerase | | Authors: | Appleby, T.C, Canales, E, Watkins, W.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1Q7I
 
 | |
1QCQ
 
 | | UBIQUITIN CONJUGATING ENZYME | | Descriptor: | PROTEIN (UBIQUITIN CONJUGATING ENZYME) | | Authors: | Cook, W.J, Jeffrey, L.C, Xu, Y, Chau, V. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4.
Biochemistry, 32, 1993
|
|
1ULA
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1VCJ
 
 | | Influenza B virus neuraminidase complexed with 1-(4-Carboxy-2-(3-pentylamino)phenyl)-5-aminomethyl-5-hydroxymethyl-pyrrolidin-2-one | | Descriptor: | 4-[(2R)-2-(AMINOMETHYL)-2-(HYDROXYMETHYL)-5-OXOPYRROLIDIN-1-YL]-3-[(1-ETHYLPROPYL)AMINO]BENZOIC ACID, NEURAMINIDASE | | Authors: | Lommer, B.S, Ali, S.M, Bajpai, S.N, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A benzoic acid inhibitor induces a novel conformational change in the active site of Influenza B virus neuraminidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OQP
 
 | |
1VSU
 
 | |
1QX2
 
 | | X-ray Structure of Calcium-loaded Calbindomodulin (A Calbindin D9k Re-engineered to Undergo a Conformational Opening) at 1.44 A Resolution | | Descriptor: | CALCIUM ION, Vitamin D-dependent calcium-binding protein, intestinal, ... | | Authors: | Bunick, C.G, Nelson, M.R, Mangahas, S, Mizoue, L.S, Bunick, G.J, Chazin, W.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-05-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Designing Sequence to Control Protein Function in an EF-Hand Protein
J.Am.Chem.Soc., 126, 2004
|
|
4EOZ
 
 | |
1QMS
 
 | | Head-to-Tail Dimer of Calicheamicin gamma-1-I Oligosaccharide Bound to DNA Duplex, NMR, 9 Structures | | Descriptor: | CALICHEAMICIN GAMMA-1-OLIGOSACCHARIDE, DNA (5'-D(*GP*CP*AP*CP*CP*TP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*AP*GP*GP*TP*GP*C)-3'), ... | | Authors: | Bifulco, G, Galeone, A, Nicolaou, K.C, Chazin, W.J, Gomez-Paloma, L. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Complex between the Head-to-Tail Dimer of Calicheamicin Gamma-1-I Oligosaccharide and a DNA Duplex Containing D(ACCT) and D(TCCT) High-Affinity Binding Sites
J.Am.Chem.Soc., 120, 1998
|
|
1R4Z
 
 | | Bacillus subtilis lipase A with covalently bound Rc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4R)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
5SBM
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z1267885772 | | Descriptor: | (3S)-3-methyl-1-(6-methylpyridin-2-yl)piperazine, 1,2-ETHANEDIOL, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBN
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z57040482 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.181 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBL
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z126932614 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBK
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z1258992717 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBO
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z2856434878 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3,4-dimethoxyphenyl)methyl]morpholine, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SBP
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z1229798311 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-6-fluorobenzoic acid, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.275 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
