3T36
 
 | |
3L32
 
 | |
6J3G
 
 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutathione S-transferase, ... | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
6J3H
 
 | | Crystal structure of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
3SI6
 
 | | RB69 DNA Polymerase Triple Mutant (L561A/S565G/Y567A) Ternary Complex with dUpNpp and a Deoxy-terminated Primer in the presence of Mg2+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-06-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SQ4
 
 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SQ1
 
 | | RB69 DNA Polymerase Ternary Complex with dUpCpp Opposite dA | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
6J3F
 
 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6JJC
 
 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP
To Be Published
|
|
6JLC
 
 | |
3SQ2
 
 | | RB69 DNA Polymerase Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
6JJA
 
 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP | | Descriptor: | CALCIUM ION, Nucleocapsid protein CP17 | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP
To Be Published
|
|
3SPY
 
 | | RB69 DNA Polymerase(L415A/L561A/S565G/Y567A) Ternary Complex with dUpCpp Opposite dA | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-04 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
3SUQ
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (AT rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
6JKI
 
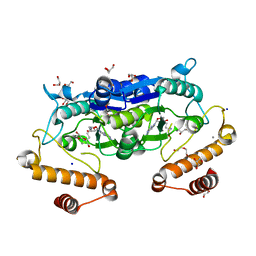 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
3SUP
 
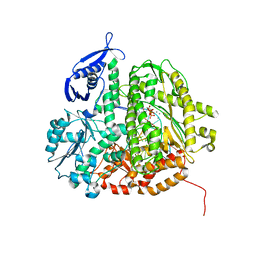 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (GC rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3L6H
 
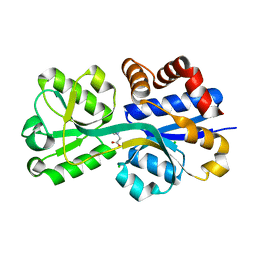 | | Crystal structure of lactococcal OpuAC in its closed-liganded conformation complexed with glycine betaine | | Descriptor: | Betaine ABC transporter permease and substrate binding protein, CHLORIDE ION, TRIMETHYL GLYCINE | | Authors: | Berntsson, R.P.A, Wolters, J.C, Gul, N, Karasawa, A, Thunnissen, A.M.W.H, Slotboom, D.J, Poolman, B. | | Deposit date: | 2009-12-23 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA.
Plos One, 5, 2010
|
|
7MZK
 
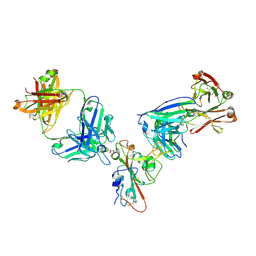 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 96 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZG
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PDI 42 heavy chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZJ
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 93 | | Descriptor: | GLYCEROL, PDI 93 heavy chain, PDI 93 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZH
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 119 | | Descriptor: | Spike protein S1, WCSL 119 heavy chain, WCSL 119 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZF
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 37 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZM
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 215 | | Descriptor: | ISOPROPYL ALCOHOL, PDI 215 heavy chain, PDI 215 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZL
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 210 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 210 heavy chain, PDI 210 light chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZI
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 | | Descriptor: | GLYCEROL, Spike protein S1, TETRAETHYLENE GLYCOL, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
