2VU1
 
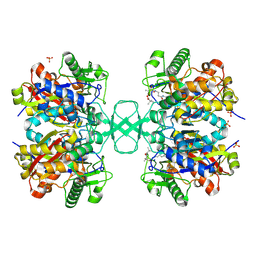 | | Biosynthetic thiolase from Z. ramigera. Complex of with O-pantheteine- 11-pivalate. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, PANTOTHENYL-AMINOETHANOL-11-PIVALIC ACID, SODIUM ION, ... | | Authors: | Kursula, P, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
2WGT
 
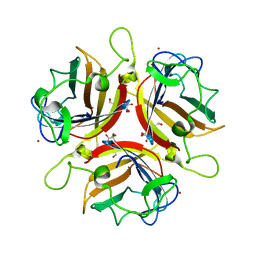 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N-propaonyl-3,5-dideoxy-D- glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
1A3W
 
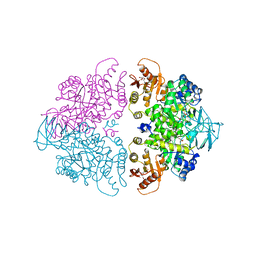 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH FBP, PG, MN2+ AND K+ | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
2C1Z
 
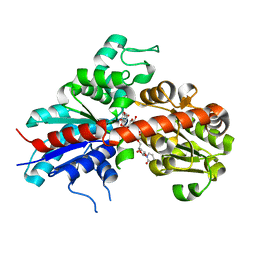 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2C1X
 
 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2BUN
 
 | | Solution structure of the BLUF domain of AppA 5-125 | | Descriptor: | APPA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Grinstead, J.S, Hsu, S.-T, Laan, W, Bonvin, A.M.J.J, Hellingwerf, K.J, Boelens, R, Kaptein, R. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling.
Chembiochem, 7, 2006
|
|
2CDX
 
 | |
2VSI
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, 4-AMINO-1-{5-O-[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]-ALPHA-D-ARABINOFURANOSYL}PYRIMIDIN-2(1H)-ONE, CALCIUM ION | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4GGC
 
 | |
2VPP
 
 | | Drosophila melanogaster deoxyribonucleoside kinase successfully activates gemcitabine in transduced cancer cell lines | | Descriptor: | DEOXYNUCLEOSIDE KINASE, GEMCITABINE, SULFATE ION | | Authors: | Knecht, W, Mikkelsen, N.E, Clausen, A, Willer, M, Gojkovic, Z, Piskur, J. | | Deposit date: | 2008-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Drosophila Melanogaster Deoxyribonucleoside Kinase Activates Gemcitabine.
Biochem.Biophys.Res.Commun., 382, 2009
|
|
2W4F
 
 | | CRYSTAL STRUCTURE OF THE FIRST PDZ DOMAIN OF HUMAN SCRIB1 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN LAP4 | | Authors: | Hozjan, V, Pilka, E.S, Roos, A.K, W Yue, W, Phillips, C, Bray, J, Cooper, C, Salah, E, Elkins, J.M, Muniz, J.R.C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Bountra, C, Doyle, D.A, Oppermann, U. | | Deposit date: | 2008-11-25 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the First Pdz Domain of Human Scrib1
To be Published
|
|
2C8W
 
 | | thrombin inhibitors | | Descriptor: | (2S,3R)-N-[5-CHLORO-2-(2,3-DIHYDRO-1H-TETRAZOL-1-YL)BENZYL]-3-HYDROXY-4-{[(4-METHOXYPHENYL)SULFONYL]AMINO}-1-PHENYLBUTA N-2-AMINIUM, HIRUDIN VARIANT-2, SODIUM ION, ... | | Authors: | Howard, N, Abell, C, Blakemore, W, Carr, R, Chessari, G, Congreve, M, Howard, S, Jhoti, H, Murray, C.W, Seavers, L.C.A, van Montfort, R.L.M. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Application of Fragment Screening and Fragment Linking to the Discovery of Novel Thrombin Inhibitors
J.Med.Chem., 49, 2006
|
|
2CD2
 
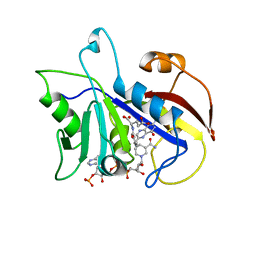 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J, Pangborn, W, Queener, S. | | Deposit date: | 1999-03-15 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
2VRE
 
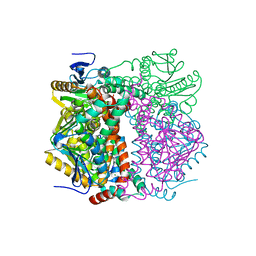 | | crystal structure of human peroxisomal delta3,5,delta2,4-dienoyl coa isomerase | | Descriptor: | CHLORIDE ION, DELTA(3,5)-DELTA(2,4)-DIENOYL-COA ISOMERASE | | Authors: | Yue, W, Guo, K, von Delft, F, Pilka, E, Murray, J, Roos, A, Kochan, G, Bountra, C, Arrowsmith, C, Wikstrom, M, Edwards, A, Oppermann, U. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peroxisomal Delta3,5, Delta2,4-Dienoyl Coa Isomerase (Ech1)
To be Published
|
|
1B47
 
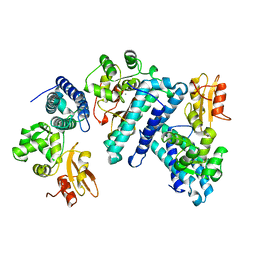 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE IN ZAP-70 | | Descriptor: | CALCIUM ION, CBL | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1999-01-06 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
2E22
 
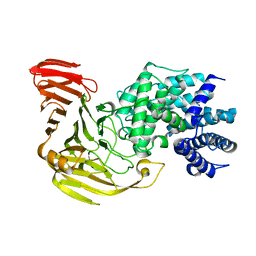 | | Crystal structure of xanthan lyase in complex with mannose | | Descriptor: | Xanthan lyase, alpha-D-mannopyranose | | Authors: | Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Structural Factor Responsible for Substrate Recognition by Bacillus sp. GL1 Xanthan Lyase that Acts Specifically on Pyruvated Side Chains of Xanthan
Biochemistry, 46, 2007
|
|
1FM1
 
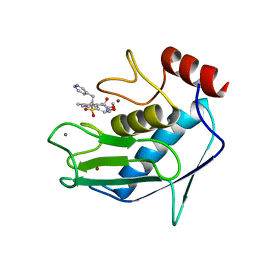 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
1AVG
 
 | |
2E5S
 
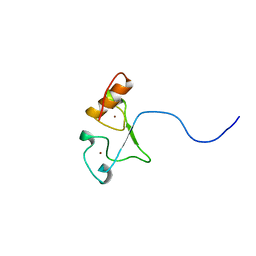 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
1BA4
 
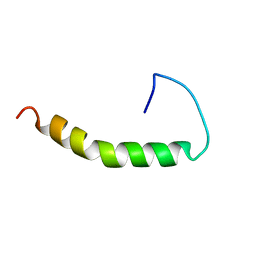 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
2E72
 
 | | Solution structure of the zinc finger domain of human KIAA0461 | | Descriptor: | Pogo transposable element with ZNF domain, ZINC ION | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-05 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger domain of human KIAA0461
To be Published
|
|
2VU2
 
 | | Biosynthetic thiolase from Z. ramigera. Complex with S-pantetheine-11- pivalate. | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-({3-oxo-3-[(2-sulfanylethyl)amino]propyl}amino)butyl 2,2-dimethylpropanoate, ACETYL-COA ACETYLTRANSFERASE, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
1BHZ
 
 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF HEN EGG WHITE LYSOZYME FROM MASC DATA | | Descriptor: | LYSOZYME | | Authors: | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2VXS
 
 | | Structure of IL-17A in complex with a potent, fully human neutralising antibody | | Descriptor: | FAB FRAGMENT, INTERLEUKIN-17A, SULFATE ION | | Authors: | Gerhardt, S, Hargreaves, D, Pauptit, R.A, Davies, R.A, Russell, C, Welsh, F, Tuske, S.J, Coales, S.J, Hamuro, Y, Needham, M.R.C, Langham, C, Barker, W, Bell, P, Aziz, A, Smith, M.J, Dawson, S, Abbott, W.M. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
J.Mol.Biol., 394, 2009
|
|
