8SFW
 
 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Arriaza, R.H, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with quercetin
To Be Published
|
|
1OSG
 
 | | Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold | | Descriptor: | BR3 derived PEPTIDE, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
4CYW
 
 | | Structure of the A_mallard_Sweden_51_2002 H10 Avian Haemmaglutinin in complex with human receptor analog 6-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Martin, S.R, Haire, L.F, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-04-15 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Receptor Binding by H10 Influenza Viruses.
Nature, 511, 2014
|
|
8PDG
 
 | | The phosphatase and C2 domains of SHIP1 with covalent Z2738285202 | | Descriptor: | DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1, ~{N}-(8-chloranylquinolin-2-yl)propanamide | | Authors: | Bradshaw, W.J, Moreira, T, Scacioc, A, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
1BS9
 
 | | ACETYLXYLAN ESTERASE FROM P. PURPUROGENUM REFINED AT 1.10 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6DOD
 
 | |
6DOP
 
 | |
1BTM
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Delboni, L.F, Mande, S.C, Hol, W.G.J. | | Deposit date: | 1995-11-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci., 4, 1995
|
|
3N2B
 
 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
4Q1S
 
 | | Yeast 20S proteasome in Complex with Kendomycin | | Descriptor: | (5R,6R,7S,8R,9R,12S,13E,16S,18S,19R,20aR)-4,7,19-trihydroxy-2,6,8,12,14,16,18-heptamethyl-6,7,8,9,10,11,12,15,16,17,18,19,20,20a-tetradecahydro-1,19:5,9-diepoxybenzo[18]annulen-3(5H)-one, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Beck, P, Heinemeyer, W, Spaeth, A, Elnakady, Y, Mueller, R, Groll, M. | | Deposit date: | 2014-04-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of the natural product kendomycin and the 20S proteasome.
J.Mol.Biol., 426, 2014
|
|
6DP3
 
 | |
4CR4
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6DPK
 
 | |
3TFT
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, pre-reaction complex with a 3,6-dihydropyrid-2-one heterocycle inhibitor | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DIMETHYL SULFOXIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Geders, T.W, Finzel, B.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism-based Inactivation by Aromatization of the Transaminase BioA Involved in Biotin Biosynthesis in Mycobaterium tuberculosis.
J.Am.Chem.Soc., 133, 2011
|
|
3N3W
 
 | | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni
TO BE PUBLISHED
|
|
8SGG
 
 | | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137D09 Fab heavy chain, Cy137D09 Fab light chain, GLYCEROL | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine
To Be Published
|
|
4AGC
 
 | | CRYSTAL STRUCTURE OF VEGFR2 (JUXTAMEMBRANE AND KINASE DOMAINS) IN COMPLEX WITH AXITINIB (AG-013736) (N-Methyl-2-(3-((E)-2-pyridin-2-yl- vinyl)-1H-indazol-6-ylsulfanyl)-benzamide) | | Descriptor: | AXITINIB, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | Authors: | Mctigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Kania, R.S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8A00
 
 | | Infectious mouse-adapted ME7 scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for prion strain diversity.
Nat.Chem.Biol., 19, 2023
|
|
8T6P
 
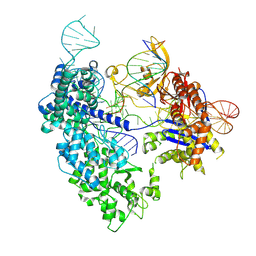 | | SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 2 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-16 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8SPQ
 
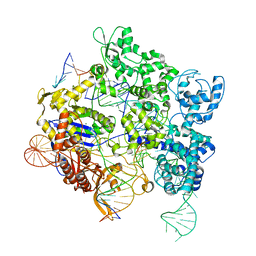 | | SpRY-Cas9:gRNA complex targeting TGG PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
4AH9
 
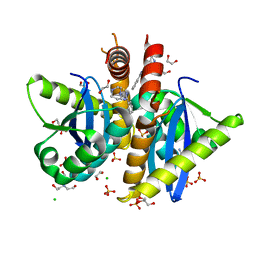 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PHENYL-1,2,4-THIADIAZOL-5-YL)-1,4-DIAZEPANE, CHLORIDE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
8T6Y
 
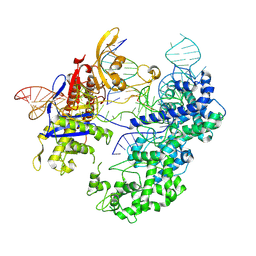 | | SpRY-Cas9:gRNA complex bound to non-target DNA with 1 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8T7S
 
 | | SpRYmer bound to NAC PAM DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
8T78
 
 | | SpRY-Cas9:gRNA complex bound to non-target DNA with 8 bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Hibshman, G.N, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
3R93
 
 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
