7GOX
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z33452106 | | Descriptor: | DIMETHYL SULFOXIDE, Protease 3C, ~{N}-(4-methyl-2-oxidanyl-phenyl)propanamide | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOY
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z375990520 | | Descriptor: | DIMETHYL SULFOXIDE, Protease 3C, methyl N-(5-methyl-1,2-oxazole-3-carbonyl)glycinate | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
1GJW
 
 | | Thermotoga maritima maltosyltransferase complex with maltose | | Descriptor: | MALTODEXTRIN GLYCOSYLTRANSFERASE, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Roujeinikova, A, Raasch, C, Burke, J, Baker, P.J, Liebl, W, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Maltosyltransferase and its Implications for the Molecular Basis of the Novel Transfer Specificity
J.Mol.Biol., 312, 2001
|
|
7GNZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1129283193 | | Descriptor: | DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)acetamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
2WE3
 
 | | EBV dUTPase inactive mutant deleted of motif V | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
7GO8
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1230013388 | | Descriptor: | 1-(oxan-4-yl)-1H-pyrazole-5-carboxylic acid, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
2WE2
 
 | | EBV dUTPase double mutant Gly78Asp-Asp131Ser with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, SULFATE ION | | Authors: | Freeman, L, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Flexible Motif V of Epstein-Barr Virus Deoxyuridine 5'-Triphosphate Pyrophosphatase is Essential for Catalysis.
J.Biol.Chem., 284, 2009
|
|
7GOC
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1318110042 | | Descriptor: | 4-{[(1M)-buta-1,2-dien-1-yl](methyl)amino}-1lambda~6~-thiane-1,1-dione, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOV
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z30904160 | | Descriptor: | DIMETHYL SULFOXIDE, N-(5-fluoro-2-methylphenyl)pyridine-4-carboxamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
2WEI
 
 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
7GP2
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z425757818 | | Descriptor: | 1-(methanesulfonyl)piperidin-4-ol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GP6
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z53116498 | | Descriptor: | 3-(2-methyl-1H-benzimidazol-1-yl)propanamide, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GP9
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z53860899 | | Descriptor: | DIMETHYL SULFOXIDE, Protease 3C, ~{N}-(1,3-benzodioxol-5-ylmethyl)ethanesulfonamide | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
6D7F
 
 | | Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide | | Descriptor: | Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
7GPP
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with POB0050 | | Descriptor: | DIMETHYL SULFOXIDE, Protease 3C, methyl (5S)-1-acetyl-5-methyl-L-prolinate | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQ5
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z2301685688 | | Descriptor: | 1H-imidazole-5-carbonitrile, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQA
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z57299966 | | Descriptor: | (1-methyl-1H-benzimidazol-2-yl)methanol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
3TYR
 
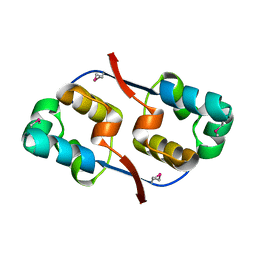 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
1FXH
 
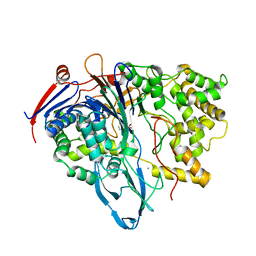 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
3QBD
 
 | |
7GQJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z667925512 | | Descriptor: | 5-METHYL-3-PHENYL-1H-PYRAZOLE, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOD
 
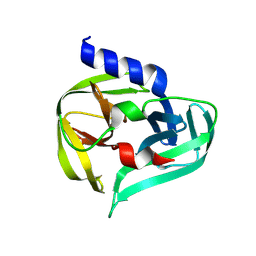 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1331830630 | | Descriptor: | 2-ethyl-N-(2-hydroxyphenyl)butanamide, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1685106505 | | Descriptor: | (3M)-5-cyclobutyl-3-(1-methyl-1H-imidazol-2-yl)-1,2,4-oxadiazole, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GQM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z362043378 | | Descriptor: | 2-phenylpyrimidin-5-ol, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
7GOL
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z1827602749 | | Descriptor: | (2S,3R)-3-methyl-N-(1,2,3-thiadiazol-5-yl)oxolane-2-carboxamide (non-preferred name), DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
