1OJ4
 
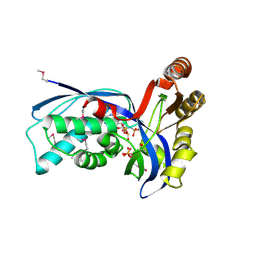 | | Ternary complex of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL KINASE, CHLORIDE ION, ... | | Authors: | Miallau, L, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2003-06-30 | | Release date: | 2003-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Isoprenoids: Crystal Structure of 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8SW1
 
 | |
3VIJ
 
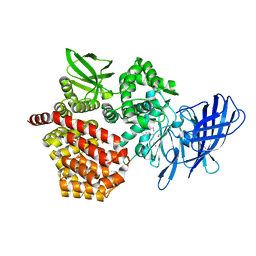
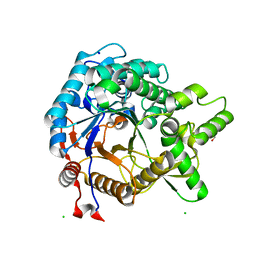 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with glucose | | Descriptor: | Beta-glucosidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A6N
 
 | | STRUCTURE OF THE TETRACYCLINE DEGRADING MONOOXYGENASE TETX IN COMPLEX WITH TIGECYCLINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2011-11-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative Dioxygen-Binding Sites and Recognition of Tigecycline and Minocycline in the Tetracycline-Degrading Monooxygenase Tetx
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VJ8
 
 | |
3NIK
 
 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
4MWA
 
 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3N8H
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis | | Descriptor: | ACETIC ACID, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis
To be Published, 2010
|
|
1G0M
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152I | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1FVT
 
 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
6DOC
 
 | |
6DOO
 
 | |
3NKV
 
 | | Crystal structure of Rab1b covalently modified with AMP at Y77 | | Descriptor: | ADENOSINE MONOPHOSPHATE, BARIUM ION, MAGNESIUM ION, ... | | Authors: | Mueller, M.P, Peters, H, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
Science, 329, 2010
|
|
7DO1
 
 | |
8STY
 
 | |
7D11
 
 | |
8T0G
 
 | |
7KWK
 
 | |
8T0H
 
 | |
8STZ
 
 | |
8T0I
 
 | |
6DEW
 
 | | Structure of human COQ9 protein with bound isoprene. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ... | | Authors: | Bingman, C.A, Lohman, D.C, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2018-05-13 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis.
Mol.Cell, 73, 2019
|
|
2VA6
 
 | | X-ray crystal structure of beta secretase complexed with compound 24 | | Descriptor: | (6S)-2-amino-6-(3'-methoxybiphenyl-3-yl)-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, BETA SECRETASE 1, IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L.L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
8TJ2
 
 | | CryoEM structure of Myxococcus xanthus type IV pilus | | Descriptor: | Type IV major pilin protein PilA | | Authors: | Zheng, W, Egelman, E.H. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tight-packing of large pilin subunits provides distinct structural and mechanical properties for the Myxococcus xanthus type IVa pilus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1FK2
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
