7RDU
 
 | |
6A0H
 
 | |
6ZZB
 
 | |
3DCL
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
6A1Z
 
 | | Crystal Structure of dimeric Kinesin-3 KIF13B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin family member 13B, MAGNESIUM ION | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-06-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2QUW
 
 | |
7RPU
 
 | |
4ZFK
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC with glutamine | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase EgtC, GLUTAMINE | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
4ZGC
 
 | | Crystal Structure Analysis of Kelch protein (with disulfide bond) from Plasmodium falciparum | | Descriptor: | Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-04-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of kelch protein with disulfide bond from Plasmodium falciparum.
to be published
|
|
7RPT
 
 | |
9B6E
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 | | Descriptor: | 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
9B6F
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850 | | Descriptor: | (8R)-8-[4-(trifluoromethyl)phenyl]-N-[(2S)-1,1,1-trifluoropropan-2-yl]-5,8-dihydro-1,7-naphthyridine-7(6H)-carboxamide, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
9B6G
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, N-(3-aminopropyl)-2-[(3-methylphenyl)methoxy]-N-[(thiophen-2-yl)methyl]benzamide, ... | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
9B6H
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
7SAN
 
 | | Crystal structure of human hypoxanthine guanine phosphoribzosyltransferase in complex with (4S,7S)-7-hydroxy-4-((guanin-9-yl)methyl)-2,5-dioxaheptan-1,7-diphosphonate | | Descriptor: | ({(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2S)-2-hydroxy-2-phosphonoethoxy]propoxy}methyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58155513 Å) | | Cite: | Stereo-Defined Acyclic Nucleoside Phosphonates are Selective and Potent Inhibitors of Parasite 6-Oxopurine Phosphoribosyltransferases.
J.Med.Chem., 65, 2022
|
|
1R0C
 
 | |
3GSD
 
 | | 2.05 Angstrom structure of a divalent-cation tolerance protein (CutA) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Structure of a Divalent-cation Tolerance Protein (CutA) from Yersinia pestis
TO BE PUBLISHED
|
|
1DII
 
 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
7UQA
 
 | | Crystal structure of the small Ultra-Red Fluorescent Protein (smURFP) | | Descriptor: | CHLORIDE ION, SODIUM ION, small Ultra-Red Fluorescent Protein (smURFP) | | Authors: | Maiti, A, Buffalo, C.Z, Saurabh, S, Montecinos-Franjola, F, Hachey, J.S, Conlon, W.J, Tran, G.N, Drobizhev, M, Moerner, W.E, Ghosh, P, Matsuo, H, Tsien, R.Y, Lin, J.Y, Rodriguez, E.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural and photophysical characterization of the small ultra-red fluorescent protein.
Nat Commun, 14, 2023
|
|
7D9Q
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 7 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(3-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9P
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 12 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(2-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9O
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 2 | | Descriptor: | (2R)-2-[[4-fluoranyl-1-[(4-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
4BDH
 
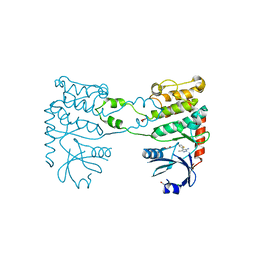 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYL-4-(THIOPHEN-2-YL)-1H-PYRAZOL-5-AMINE, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
7H6V
 
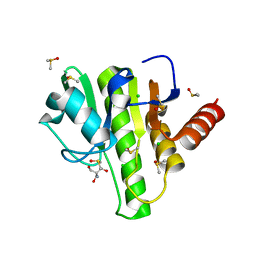 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1262633486 (CHIKV_MacB-x0394) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(difluoromethyl)-1-methyl-1H-pyrazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7SBB
 
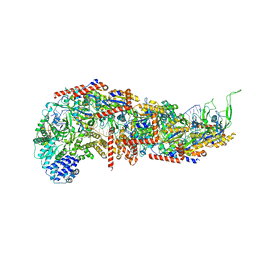 | |
