6P49
 
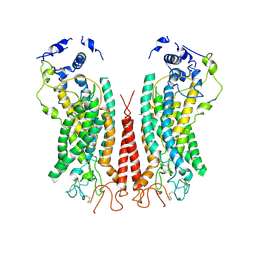 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl2 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
8CLI
 
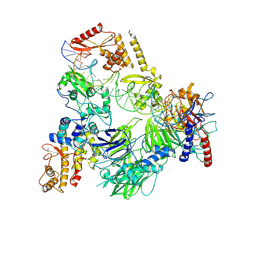 | | TFIIIC TauB-DNA monomer | | Descriptor: | DNA (35-MER), General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
6P7K
 
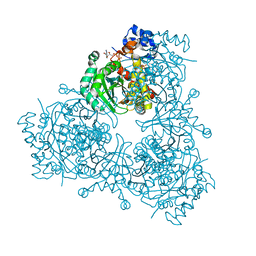 | | Structure of HMG-CoA reductase from Burkholderia cenocepacia | | Descriptor: | 3-hydroxy-3-methylglutaryl coenzyme A reductase, ADENOSINE-5'-DIPHOSPHATE, COENZYME A | | Authors: | Walker, A.M, Peacock, R.B, Hicks, C.W, Dewing, S.M, Lewis, K.M, Abboud, J, Stewart, S.W.A, Kang, C, Watson, J.M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Structural and Functional Characterization of Dynamic Oligomerization in Burkholderia cenocepacia HMG-CoA Reductase.
Biochemistry, 58, 2019
|
|
8CLK
 
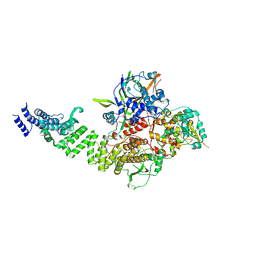 | | TFIIIC TauA complex | | Descriptor: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 3, General transcription factor 3C polypeptide 5, ... | | Authors: | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
1F6Y
 
 | |
4CVH
 
 | | Crystal structure of human isoprenoid synthase domain-containing protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPRENOID SYNTHASE DOMAIN-CONTAINING PROTEIN, ... | | Authors: | Kopec, J, Froese, D.S, Krojer, T, Newman, J, Kiyani, W, Goubin, S, Strain-Damerell, C, Vollmar, M, von Delft, F, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Lefeber, D.J, Yue, W.W. | | Deposit date: | 2014-03-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Human Ispd is a Cytidyltransferase Required for Dystroglycan O-Mannosylation.
Chem.Biol., 22, 2015
|
|
2GTT
 
 | | Crystal structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H. | | Deposit date: | 2006-04-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structure of the Rabies Virus Nucleoprotein-RNA Complex
Science, 313, 2006
|
|
2HIT
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylethanolamine | | Descriptor: | (1R)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
6ISH
 
 | | Structure of 9N-I DNA polymerase incorporation with 3'-AL in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*(DAL))-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6PID
 
 | |
8TLU
 
 | | E. coli MraY mutant-T23P | | Descriptor: | Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Li, Y.E, Clemons, W.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa.
Nat Microbiol, 9, 2024
|
|
6P46
 
 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
4XOX
 
 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
6PI8
 
 | |
6PKM
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#8) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.17 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
8TW0
 
 | | Crystal Structure of a synthetic ABC heterotrimeric Collagen-like Peptide at 1.53 A | | Descriptor: | Collagen Mimetic Peptide A, Collagen Mimetic Peptide B, Collagen Mimetic Peptide C, ... | | Authors: | Miller, M.D, Cole, C.C, Xu, W, Walker, D.R, Hulgan, S.A.H, Pogostin, B.H, Swain, J.W.R, Duella, R, Misiura, M, Wang, X, Kolomeisky, A.B, Phillips Jr, G.N, Hartgerink, J.D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Heterotrimeric collagen helix with high specificity of assembly results in a rapid rate of folding.
Nat.Chem., 2024
|
|
4XXH
 
 | | TREHALOSE REPRESSOR FROM ESCHERICHIA COLI | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, HTH-type transcriptional regulator TreR | | Authors: | Hars, U, Horlacher, R, Boos, W, Smart, O.S, Bricogne, G, Welte, W, Diederichs, K. | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF THE EFFECTOR-BINDING DOMAIN OF THE TREHALOSE-REPRESSOR OF ESCHERICHIA COLI, A MEMBER OF THE LACI FAMILY, IN ITS COMPLEXES WITH INDUCER TREHALOSE-6-PHOSPHATE AND NONINDUCER TREHALOSE.
PROTEIN SCI., 7, 1998
|
|
6PKQ
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.85A resolution (#11) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6P47
 
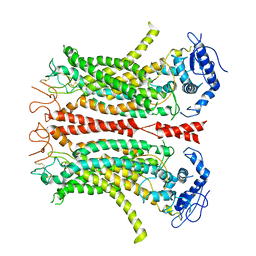 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | Descriptor: | Anoctamin-6 | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
7MXY
 
 | | Cryo-EM structure of PfFNT-inhibitor complex | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yu, E.W, Su, C, Lyu, M. | | Deposit date: | 2021-05-19 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT
EMBO Rep, 22, 2021
|
|
3FFD
 
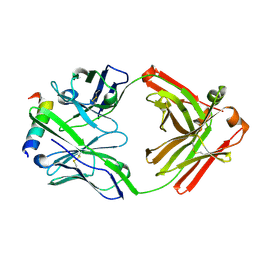 | | Structure of parathyroid hormone-related protein complexed to a neutralizing monoclonal antibody | | Descriptor: | Monoclonal antibody, heavy chain, Fab fragment, ... | | Authors: | Mckinstry, W.J, Polekhina, G, Diefenbach-Jagger, H, Ho, P.W.M, Sato, K, Onuma, E, Gillespie, M.T, Martin, T.J, Parker, M.W. | | Deposit date: | 2008-12-03 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody discrimination between two hormones that recognize the parathyroid hormone receptor
J.Biol.Chem., 284, 2009
|
|
6PRZ
 
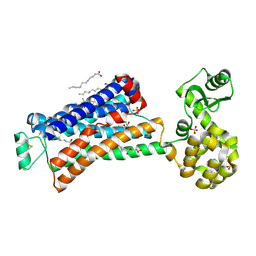 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
3NGX
 
 | |
3A7I
 
 | | Human MST3 kinase in complex with adenine | | Descriptor: | ADENINE, Serine/threonine kinase 24 (STE20 homolog, yeast) | | Authors: | Ko, T.P, Jeng, W.Y, Liu, C.I, Lai, M.D, Wang, A.H.J. | | Deposit date: | 2009-09-26 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of human MST3 kinase in complex with adenine, ADP and Mn2+.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4XZZ
 
 | | Structure of Helicobacter pylori Csd6 in the ligand-free state | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
