4A5S
 
 | | CRYSTAL STRUCTURE OF HUMAN DPP4 IN COMPLEX WITH A NOVAL HETEROCYCLIC DPP4 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3S)-3-AMINOPIPERIDIN-1-YL]-5-BENZYL-4-OXO-3-(QUINOLIN-4-YLMETHYL)-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDINE-7-CARBONITRILE, DIPEPTIDYL PEPTIDASE 4 SOLUBLE FORM, ... | | Authors: | Ostermann, N, Kroemer, M, Zink, F, Gerhartz, B, Sutton, J.M, Clark, D.E, Dunsdon, S.J, Fenton, G, Fillmore, A, Harris, N.V, Higgs, C, Hurley, C.A, Krintel, S.L, MacKenzie, R.E, Duttaroy, A, Gangl, E, Maniara, W, Sedrani, R, Namoto, K, Sirockin, F, Trappe, J, Hassiepen, U, Baeschlin, D.K. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel Heterocyclic Dpp-4 Inhibitors for the Treatment of Type 2 Diabetes.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4AAQ
 
 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
2ASN
 
 | | Crystal structure of D1A mutant of nitrophorin 2 complexed with imidazole | | Descriptor: | IMIDAZOLE, Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-23 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
4H9Q
 
 | | Complex structure 4 of DAXX(E225A)/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4YQ6
 
 | |
1E4Y
 
 | |
4HC6
 
 | | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, H.-B, Han, G.-W, Hung, L.-W, Terwilliger, C.T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution
To be Published
|
|
4YQC
 
 | |
5UHB
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.29 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
6NN5
 
 | | The structure of human liver pyruvate kinase, hLPYK-W527H | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pyruvate kinase PKLR | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1ZNQ
 
 | | Crystal Structure of Human Liver GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, liver, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ismail, S.A, Park, H.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of Human liver GAPDH
To be Published
|
|
1EAP
 
 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
4HCS
 
 | | Structure of Novel subfamily CX chemokine solved by sulfur SAD | | Descriptor: | Uncharacterized protein | | Authors: | Rajasekaran, D, Fan, C, Meng, W, Pflugrath, J.W, Lolis, E.J. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-16 | | Last modified: | 2014-04-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insight into the evolution of a new chemokine family from zebrafish.
Proteins, 82, 2014
|
|
1ED3
 
 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
1KI6
 
 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
5U2G
 
 | | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae.
To Be Published
|
|
2B3B
 
 | | Thermus thermophilus Glucose/Galactose Binding Protein With Bound Glucose | | Descriptor: | alpha-D-glucopyranose, beta-D-glucopyranose, glucose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Warren, J.J, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2005-09-20 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of a thermophilic glucose binding protein reveals adaptations that interconvert mono and di-saccharide binding sites.
J.Mol.Biol., 362, 2006
|
|
1KJW
 
 | | SH3-Guanylate Kinase Module from PSD-95 | | Descriptor: | POSTSYNAPTIC DENSITY PROTEIN 95, SULFATE ION | | Authors: | McGee, A.W, Dakoji, S.R, Olsen, O, Bredt, D.S, Lim, W.A, Prehoda, K.E. | | Deposit date: | 2001-12-05 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the SH3-Guanylate Kinase Module from PSD-95 Suggests a Mechanism for Regulated Assembly of MAGUK Scaffolding Proteins
Mol.Cell, 8, 2001
|
|
3T5P
 
 | | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | BmrU protein, MAGNESIUM ION | | Authors: | Hou, J, Zheng, H, Chruszcz, M, Cooper, D.R, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
To be Published
|
|
2B5W
 
 | | Crystal structure of D38C glucose dehydrogenase mutant from Haloferax mediterranei | | Descriptor: | CITRATE ANION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AH7
 
 | | Crystal structure of nitrophorin 2 aqua complex | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-07-27 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
6NMZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with an intentionally produced fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with an intentionally produced fragment degradation product B9D
To Be Published
|
|
6NN7
 
 | | The structure of human liver pyruvate kinase, hLPYK-GGG | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4C2Y
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA co-factor | | Descriptor: | CITRIC ACID, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE 1, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
1Z0M
 
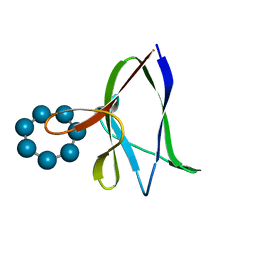 | | the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit | | Descriptor: | 5'-AMP-activated protein kinase, beta-1 subunit, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Polekhina, G, Gupta, A, van Denderen, B.J, Feil, S.C, Kemp, B.E, Stapleton, D, Parker, M.W. | | Deposit date: | 2005-03-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Glycogen Recognition by AMP-Activated Protein Kinase.
Structure, 13, 2005
|
|
