1QBA
 
 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
4W5U
 
 | |
4W5Z
 
 | |
5EKA
 
 | | HU DNA-binding protein from Thermus thermophilus | | Descriptor: | DNA-binding protein HU, GLYCEROL | | Authors: | Papageorgiou, A, Adam, P, Stavros, P, Nounesis, G, Meijers, R, Petratos, K, Vorgias, C.E. | | Deposit date: | 2015-11-03 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses.
Extremophiles, 20, 2016
|
|
1HUE
 
 | | HISTONE-LIKE PROTEIN | | Descriptor: | HU PROTEIN | | Authors: | Vis, H, Mariani, M, Vorgias, C.E, Wilson, K.S, Kaptein, R, Boelens, R. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HU protein from Bacillus stearothermophilus.
J.Mol.Biol., 254, 1995
|
|
4MB4
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB5
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1CTN
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
4HMD
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HME
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMC
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina | | Descriptor: | Chitinase 60, GLYCEROL, SODIUM ION | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-14 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1QBB
 
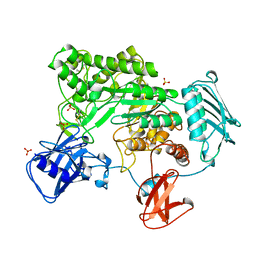 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1RIY
 
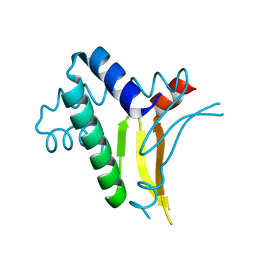 | |
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-25 | | Release date: | 2001-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1B8Z
 
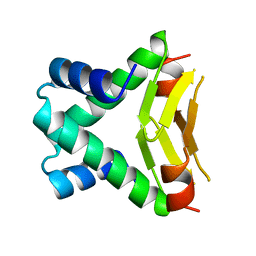 | | HU FROM THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (HISTONELIKE PROTEIN HU) | | Authors: | Christodoulou, E, Rypniewski, W.R, Vorgias, C.E. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, overproduction, purification and crystallization of the DNA binding protein HU from the hyperthermophilic eubacterium Thermotoga maritima.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
