4WR2
 
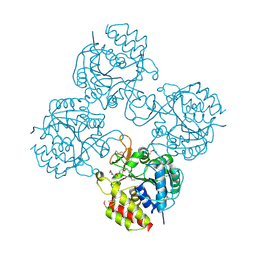 | | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Pyrimidine-specific ribonucleoside hydrolase RihA | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site
To be published
|
|
4LYY
 
 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Shewanella pealeana ATCC 700345, NYSGRC Target 029677. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Shewanella pealeana ATCC 700345, NYSGRC Target 029677.
To be Published
|
|
4MCI
 
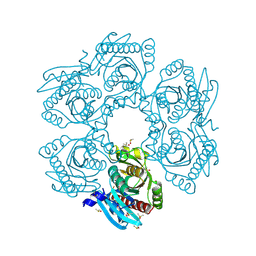 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520. | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Uridine phosphorylase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with DMSO, NYSGRC Target 029520.
To be Published
|
|
5DK6
 
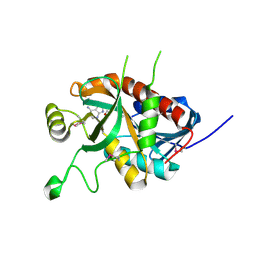 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
2LJA
 
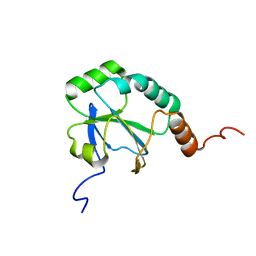 | | Solution Structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus | | Descriptor: | Putative thiol-disulfide oxidoreductase | | Authors: | Harris, R, Seidel, R.D, Hillerich, B, Gizzi, A, Kar, A, Lafleur, J, Chamala, S, Foti, R, Villegas, G, Evans, B, Zencheck, W, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-09 | | Release date: | 2011-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a putative thiol-disulfide oxidoreductase from Bacteroides vulgatus
To be Published
|
|
4RPF
 
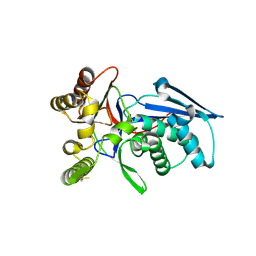 | | Crystal structure of homoserine kinase from Yersinia pestis Nepal516, NYSGRC target 032715 | | Descriptor: | CITRIC ACID, Homoserine kinase | | Authors: | Ptskovsky, Y, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Homoserine Kinase from Yersinia Pestis Nepal516, Nysgrc Target 032715
To be Published
|
|
4TYM
 
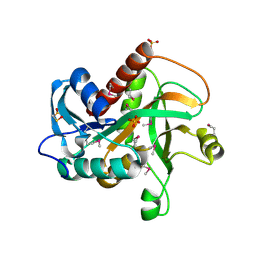 | | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Streptococcus agalactiae 2603V/R, NYSGRC Target 030935.
To Be Published
|
|
4PFQ
 
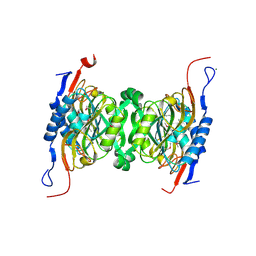 | | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 029763. | | Descriptor: | Hypoxanthine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of hypoxanthine phosphoribosyltransferase from Brachybacterium faecium DSM 4810, NYSGRC Target 0299763.
to be published
|
|
4NSN
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry | | Descriptor: | ADENINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry.
To be Published
|
|
4NS1
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
To be Published
|
|
4P52
 
 | | Crystal structure of homoserine kinase from Cytophaga hutchinsonii ATCC 33406, NYSGRC Target 032717. | | Descriptor: | Homoserine kinase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of homoserine kinase from Cytophaga hutchinsonii ATCC 33406, NYSGRC Target 032717.
to be published
|
|
4E6M
 
 | | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium)
To be Published
|
|
4F3X
 
 | | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aldehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD
To be Published
|
|
4FFU
 
 | | CRYSTAL STRUCTURE OF putative MaoC-like (monoamine oxidase-like) protein, similar to NodN from Sinorhizo Bium meliloti 1021 | | Descriptor: | CHLORIDE ION, oxidase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC-like (monoamine oxidase-like) protein, similar to NodN from Sinorhizo
Bium meliloti 1021
To be Published
|
|
4E4G
 
 | | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of putative Methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
4G2N
 
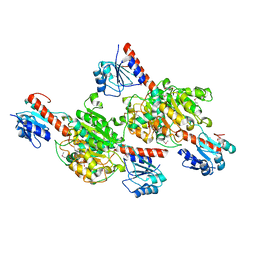 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | Descriptor: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
4HAD
 
 | | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42 | | Descriptor: | Probable oxidoreductase protein, SODIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of probable oxidoreductase protein from Rhizobium etli CFN 42
To be Published
|
|
4EZB
 
 | | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021 | | Descriptor: | uncharacterized conserved protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021
To be Published
|
|
4E3E
 
 | | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl | | Descriptor: | MaoC domain protein dehydratase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative MaoC domain protein dehydratase from Chloroflexus aurantiacus J-10-fl
To be Published
|
|
4E3A
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 | | Descriptor: | ADENOSINE, sugar kinase protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42
To be Published
|
|
4GWB
 
 | | Crystal structure of putative Peptide methionine sulfoxide reductase from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Peptide methionine sulfoxide reductase MsrA 3 | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative Peptide methionine sulfoxide reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4IYM
 
 | | Crystal structure of putative methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD, target 011934 | | Descriptor: | MAGNESIUM ION, Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-28 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD, target 011934
To be Published
|
|
4J3Z
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Jannaschia sp. CCS1 | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Jannaschia sp. CCS1
To be Published
|
|
4IX1
 
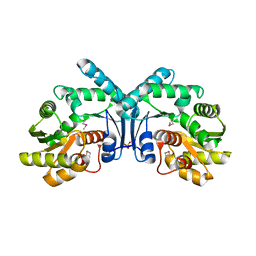 | | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205 | | Descriptor: | PHOSPHATE ION, hypothetical protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205
To be Published
|
|
4J6F
 
 | | Crystal structure of putative alcohol dehydrogenase from Sinorhizobium meliloti 1021, NYSGRC-Target 012230 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative alcohol dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alcohol dehydrogenase from Sinorhizobium meliloti 1021, NYSGRC-Target 012230
To be Published
|
|
