5HCW
 
 | |
5V0F
 
 | | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone | | Descriptor: | 4-arsanyl-2-nitrophenol, CHLORIDE ION, FE (III) ION, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2017-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone.
To Be Published
|
|
3Q5C
 
 | |
3FQ5
 
 | |
3R86
 
 | |
3E9W
 
 | |
3QK4
 
 | |
3TCI
 
 | |
3G2R
 
 | |
3G2A
 
 | |
3HS1
 
 | |
3HQE
 
 | |
3IXN
 
 | |
5C68
 
 | |
5C4P
 
 | |
5C6X
 
 | | Crystal structure of C-As lyase with Co(II) | | Descriptor: | COBALT (II) ION, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of C-As lyase with Co(II)
To Be Published
|
|
5DRH
 
 | | Crystal structure of apo C-As lyase | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of apo C-As lyase
To Be Published
|
|
5DFG
 
 | |
5D4F
 
 | | Crystal structure of C-As lyase with Fe(III) | | Descriptor: | CHLORIDE ION, FE (III) ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-08-07 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
5CB9
 
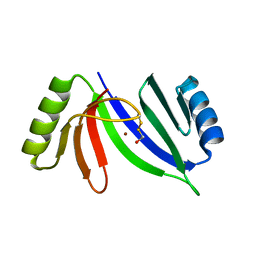 | | Crystal structure of C-As lyase with mercaptoethonal | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
6M7G
 
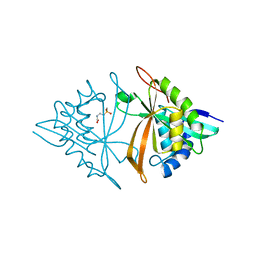 | | Crystal structure of ArsN, N-acetyltransferase with substrate phosphinothricin from Pseudomonas putida KT2440 | | Descriptor: | PHOSPHINOTHRICIN, Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
5JTF
 
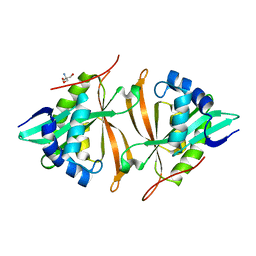 | | Crystal structure of ArsN N-acetyltransferase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
6XA0
 
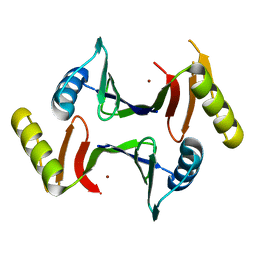 | | Crystal structure of C-As lyase with mutation K105R with Ni(II) | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, NICKEL (II) ION | | Authors: | Venkadesh, S, Yoshinaga, M, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The ArsI C-As lyase: Elucidating the catalytic mechanism of degradation of organoarsenicals.
J.Inorg.Biochem., 232, 2022
|
|
6XCK
 
 | | Crystal structure of C-As lyase with mutation K105E | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Venkadesh, S, Yoshinaga, M, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The ArsI C-As lyase: Elucidating the catalytic mechanism of degradation of organoarsenicals.
J.Inorg.Biochem., 232, 2022
|
|
5WPH
 
 | | Crystal structure of ArsN, N-acetyltransferase with substrate AST from Pseudomonas putida KT2440 | | Descriptor: | (2S)-2-amino-4-[hydroxy(methyl)arsoryl]butanoic acid, Phosphinothricin N-acetyltransferase, SODIUM ION | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2017-08-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
