2R0Y
 
 | | Structure of the Rsc4 tandem bromodomain in complex with an acetylated H3 peptide | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, Histone H3 peptide | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
5VFZ
 
 | |
7RDE
 
 | | Human Triose Phosphate Isomerase Q181P | | Descriptor: | BROMIDE ION, CALCIUM ION, Triosephosphate isomerase | | Authors: | VanDemark, A.P, Kowalski, J. | | Deposit date: | 2021-07-09 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Itavastatin and resveratrol increase triosephosphate isomerase protein in a newly identified variant of TPI deficiency.
Dis Model Mech, 15, 2022
|
|
1JAT
 
 | | Mms2/Ubc13 Ubiquitin Conjugating Enzyme Complex | | Descriptor: | Ubiquitin-Conjugating Enzyme E2-17.5 KDA, Ubiquitin-Conjugating Enzyme Variant Mms2 | | Authors: | VanDemark, A.P, Hofmann, R.M, Tsui, C, Pickart, C.M, Wolberger, C. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
1JBB
 
 | | Ubiquitin Conjugating Enzyme, Ubc13 | | Descriptor: | ubiquitin conjugating enzyme E2-17.5 KDA | | Authors: | VanDemark, A.P, Hofmann, R.M, Tsui, C, Pickart, C.M, Wolberger, C. | | Deposit date: | 2001-06-03 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
3BIP
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16 | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BIT
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FACT complex subunit SPT16, ... | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3BIQ
 
 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16, GLYCEROL | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
2GCJ
 
 | | Crystal Structure of the Pob3 middle domain | | Descriptor: | Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2GCL
 
 | | Structure of the Pob3 Middle domain | | Descriptor: | CHLORIDE ION, Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2R10
 
 | | Structure of an acetylated Rsc4 tandem bromodomain Histone Chimera | | Descriptor: | 1,2-ETHANEDIOL, Chromatin structure-remodeling complex protein RSC4, LINKER, ... | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0S
 
 | | Crystal Structure of the Rsc4 tandem bromodomain | | Descriptor: | Chromatin structure-remodeling complex protein RSC4 | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R3C
 
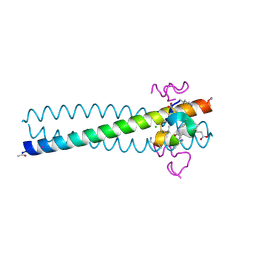 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | Descriptor: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0V
 
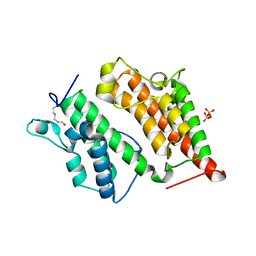 | | Structure of the Rsc4 tandem bromodomain acetylated at K25 | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, SULFATE ION | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R5B
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | HIV entry inhibitor PIE7, SULFATE ION, gp41 N-peptide | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8EXZ
 
 | |
6UIH
 
 | |
4ZVJ
 
 | | Structure of human triose phosphate isomerase K13M | | Descriptor: | POTASSIUM ION, SODIUM ION, Triosephosphate isomerase | | Authors: | Amrich, C.G, Smith, C, Heroux, A, VanDemark, A.P. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6996 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim. Biophys. Acta, 1852, 2015
|
|
6NLH
 
 | | Structure of human triose phosphate isomerase R189A | | Descriptor: | BROMIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Richards, K.R, Roland, B.P, Palladino, M.J, VanDemark, A.P. | | Deposit date: | 2019-01-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Missense variant in TPI1 (Arg189Gln) causes neurologic deficits through structural changes in the triosephosphate isomerase catalytic site and reduced enzyme levels in vivo.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
3THF
 
 | |
3V46
 
 | | Crystal Structure of Yeast Cdc73 C-Terminal Domain | | Descriptor: | Cell division control protein 73 | | Authors: | Amrich, C.G, VanDemark, A.P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin.
J.Biol.Chem., 287, 2012
|
|
5E8B
 
 | |
4POD
 
 | | Structure of Triosephosphate Isomerase I170V mutant human enzyme. | | Descriptor: | BROMIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Amrich, C.G, Aslam, A.A, Heroux, A, VanDemark, A.P. | | Deposit date: | 2014-02-25 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim.Biophys.Acta, 1852, 2015
|
|
5EMX
 
 | |
