8STG
 
 | |
2QND
 
 | |
8SWE
 
 | | FGFR2 Kinase Domain Bound to Reversible Inhibitor Cmpd 3 | | Descriptor: | Fibroblast growth factor receptor 2, GLUTATHIONE, GLYCEROL, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TU6
 
 | | CryoEM structure of PI3Kalpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Valverde, R, Shi, H, Holliday, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8U1F
 
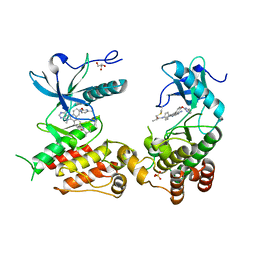 | | FGFR2 Kinase Domain Bound to Irreversible Inhibitor Cmpd 10 | | Descriptor: | Fibroblast growth factor receptor 2, GLYCEROL, N-[4-(4-amino-7-methyl-5-{4-[(4-methylpyrimidin-2-yl)oxy]phenyl}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)phenyl]-2-methylpropanamide, ... | | Authors: | Valverde, R, Foster, L. | | Deposit date: | 2023-08-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5T6J
 
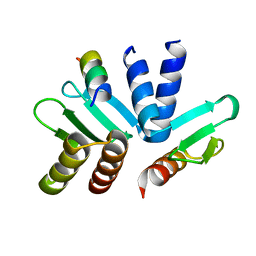 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | Kinetochore protein SPC24, Kinetochore protein SPC25, Kinetochore-associated protein DSN1 | | Authors: | Valverde, R, Jenni, S, Dimitrova, Y, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5TD8
 
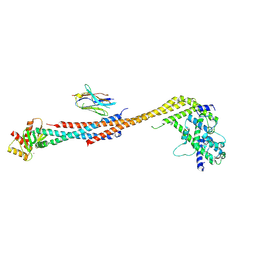 | | Crystal structure of an Extended Dwarf Ndc80 Complex | | Descriptor: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | Authors: | Valverde, R, Ingram, J, Harrison, S.C. | | Deposit date: | 2016-09-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.531 Å) | | Cite: | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
5TCS
 
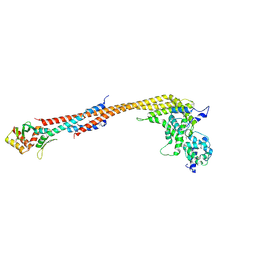 | | Crystal structure of a Dwarf Ndc80 Tetramer | | Descriptor: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | Authors: | Valverde, R, Harrison, S.C. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8313 Å) | | Cite: | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
3FWV
 
 | | Crystal Structure of a Redesigned TPR Protein, T-MOD(VMY), in Complex with MEEVF Peptide | | Descriptor: | Heat shock protein HSP 90-beta, Hsc70/Hsp90-organizing protein, NICKEL (II) ION | | Authors: | Jackrel, M.E, Valverde, R, Regan, L. | | Deposit date: | 2009-01-19 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redesign of a protein-peptide interaction: characterization and applications
Protein Sci., 18, 2009
|
|
8TSA
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 2 | | Descriptor: | 5-(3-bromo-5-fluorobenzamido)-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J, Valverde, R. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
5T51
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0E05809p, KLLA0F02343p, SULFATE ION | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2007 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5T59
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, KLLA0B13629p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5T58
 
 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0C15939p, KLLA0D15741p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2131 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
