3VNR
 
 | |
3UES
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3VNS
 
 | |
3VNQ
 
 | |
3UET
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3WID
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with NADP | | Descriptor: | Glucose 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WIE
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3STL
 
 | | KcsA potassium channel mutant Y82C with Cadmium bound | | Descriptor: | CADMIUM ION, POTASSIUM ION, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
3STZ
 
 | | KcsA potassium channel mutant Y82C with nitroxide spin label | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
3VTF
 
 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WRV
 
 | |
3WB1
 
 | | HcgB from Methanocaldococcus jannaschii | | Descriptor: | SULFATE ION, Uncharacterized protein MJ0488 | | Authors: | Tamura, H, Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2013-05-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the HcgB enzyme in [Fe]-hydrogenase-cofactor biosynthesis.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3WRY
 
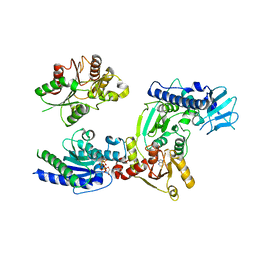 | | Crystal structure of helicase complex 2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZA0
 
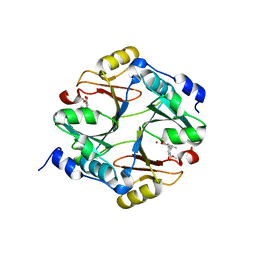 | | Crystal structure of mouse glyoxalase I complexed with methyl-gerfelin | | Descriptor: | Glyoxalase I, ZINC ION, methyl 4-(2,3-dihydroxy-5-methylphenoxy)-2-hydroxy-6-methylbenzoate | | Authors: | Okumura, H, Kawatani, M, Osada, H. | | Deposit date: | 2007-09-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZC0
 
 | | Crystal structure of an archaeal alanine:glyoxylate aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Alanine glyoxylate transaminase, IMIDAZOLE, ... | | Authors: | Sakuraba, H, Yoneda, K, Tsuge, H, Ohshima, T. | | Deposit date: | 2007-10-31 | | Release date: | 2008-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an archaeal alanine:glyoxylate aminotransferase
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZHP
 
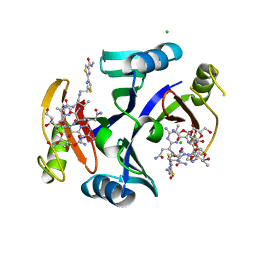 | | Crystal structure of bleomycin-binding protein from Streptoalloteichus hindustanus complexed with bleomycin derivative | | Descriptor: | Bleomycin resistance protein, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | Deposit date: | 2008-02-06 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Affinity Relationship Study of Bleomycins and Shble protein Using a Chemical Array
To be Published
|
|
2ZVR
 
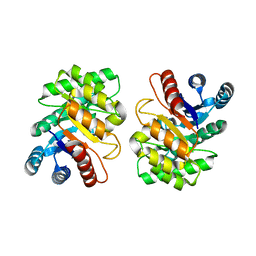 | |
2Z5G
 
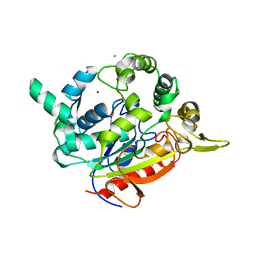 | | Crystal structure of T1 lipase F16L mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermostable lipase, ... | | Authors: | Matsumura, H, Yamamoto, T, Inoue, T, Kai, Y. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
1D8J
 
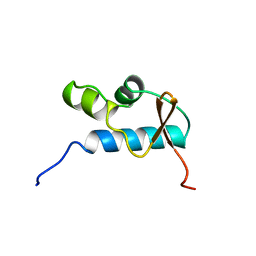 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1UAN
 
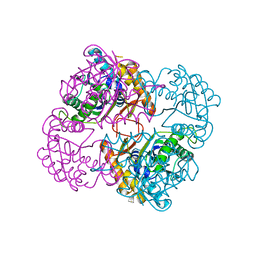 | | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1542 | | Authors: | Handa, N, Terada, T, Tame, J.R.H, Park, S.-Y, Kinoshita, K, Ota, M, Nakamura, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8
PROTEIN SCI., 12, 2003
|
|
2DC0
 
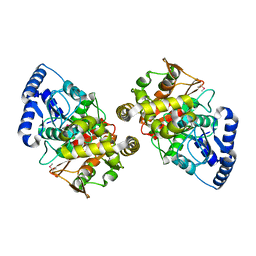 | | Crystal structure of amidase | | Descriptor: | probable amidase | | Authors: | Ohshima, T, Sakuraba, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Satoh, S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-17 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amidase
To be Published
|
|
1D06
 
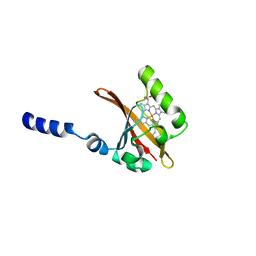 | | STRUCTURAL BASIS OF DIMERIZATION AND SENSORY MECHANISMS OF OXYGEN-SENSING DOMAIN OF RHIZOBIUM MELILOTI FIXL DETERMINED AT 1.4A RESOLUTION | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrogen fixation regulatory protein fixL | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 1999-09-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
5X9S
 
 | | Crystal structure of fully modified H-Ras-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Ke, H, Murashima, Y, Taniguchi-Tamura, H, Miyamoto, R, Yoshikawa, Y, Kumasaka, T, Mizohata, E, Edamatsu, H, Kataoka, T. | | Deposit date: | 2017-03-09 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for intramolecular interaction of post-translationally modified H-RasGTP prepared by protein ligation
FEBS Lett., 591, 2017
|
|
3AGV
 
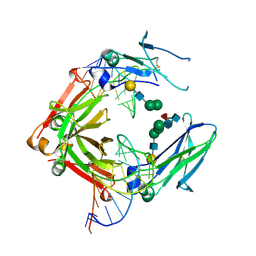 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
5B37
 
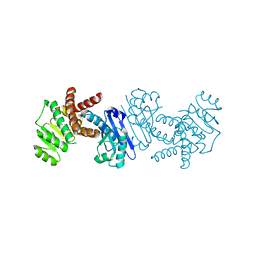 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
