6KXM
 
 | |
6KST
 
 | |
6KXN
 
 | | Crystal structure of W50A mutant of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
6KXL
 
 | | Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose | | Descriptor: | 1,2-ETHANEDIOL, 2-METHOXYETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ueda, M, Shimosaka, M, Arai, R. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of CsChiL, a chitinase from Chitiniphilus shinanonensis
To be published
|
|
6M4K
 
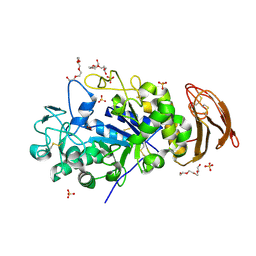 | | X-ray crystal structure of wild type alpha-amylase I from Eisenia fetida | | Descriptor: | ACETATE ION, Alpha-amylase, CALCIUM ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M4M
 
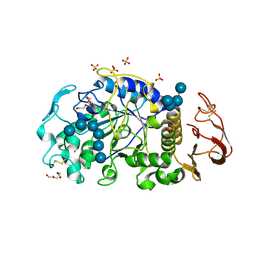 | | X-ray crystal structure of the E249Q mutan of alpha-amylase I and maltohexaose complex from Eisenia fetida | | Descriptor: | Alpha-amylase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M4L
 
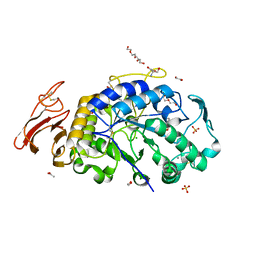 | | X-ray crystal structure of the E249Q mutant of alpha-amylase I from Eisenia fetida | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8IHW
 
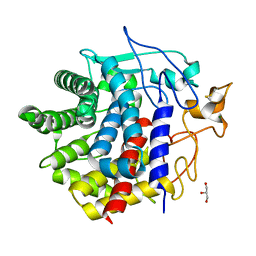 | | X-ray crystal structure of D43R mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHX
 
 | | X-ray crystal structure of N372D mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHY
 
 | | X-ray crystal structure of Q387E mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
1WPX
 
 | | Crystal structure of carboxypeptidase Y inhibitor complexed with the cognate proteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase Y, Carboxypeptidase Y inhibitor, ... | | Authors: | Mima, J, Hayashida, M, Fujii, T, Narita, Y, Hayashi, R, Ueda, M, Hata, Y. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the carboxypeptidase y inhibitor i(c) in complex with the cognate proteinase reveals a novel mode of the proteinase-protein inhibitor interaction
J.Mol.Biol., 346, 2005
|
|
5Y6T
 
 | | Crystal structure of endo-1,4-beta-mannanase from Eisenia fetida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, endo-1,4-beta-mannanase | | Authors: | Hirano, Y, Ueda, M, Tamada, T. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gene cloning, expression, and X-ray crystallographic analysis of a beta-mannanase from Eisenia fetida.
Enzyme.Microb.Technol., 117, 2018
|
|
5Z39
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum form II | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
5Z1N
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
3W6E
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6D
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6B
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 | | Descriptor: | GLYCEROL, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6F
 
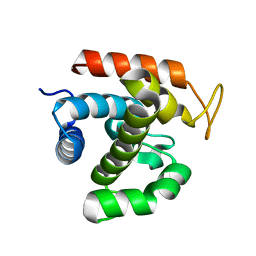 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6C
 
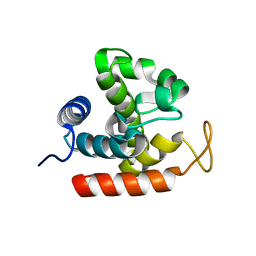 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3X39
 
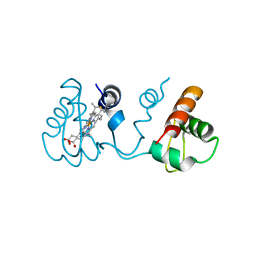 | | Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551 | | Descriptor: | Cytochrome c-551, HEME C | | Authors: | Nagao, S, Ueda, M, Osuka, H, Komori, H, Kamikubo, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Plos One, 10, 2015
|
|
1O82
 
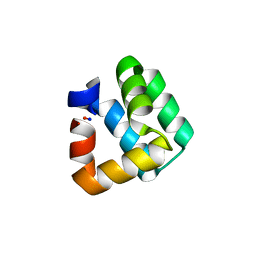 | | X-RAY STRUCTURE OF BACTERIOCIN AS-48 AT PH 4.5. SULPHATE BOUND FORM | | Descriptor: | GLYCEROL, PEPTIDE ANTIBIOTIC AS-48, SULFATE ION | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Galvez, A, Martinez-Bueno, M, Maqueda, M, Cruz, V, Albert, A. | | Deposit date: | 2002-11-22 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of Bacteriocin as-48: From Soluble State to Membrane Bound State
J.Mol.Biol., 334, 2003
|
|
1O84
 
 | | Crystal Structure of Bacteriocin AS-48. N-decyl-beta-D-maltoside Bound. | | Descriptor: | DECANE, GLYCEROL, PEPTIDE ANTIBIOTIC AS-48, ... | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Galvez, A, Valdivia, E, Maqueda, M, Cruz, V, Albert, A. | | Deposit date: | 2002-11-25 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Bacteriocin as-48: From Soluble State to Membrane Bound State
J.Mol.Biol., 334, 2003
|
|
1O83
 
 | | Crystal Structure of Bacteriocin AS-48 at pH 7.5, phosphate bound. Crystal form I | | Descriptor: | GLYCEROL, PEPTIDE ANTIBIOTIC AS-48, PHOSPHATE ION | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Galvez, A, Valdivia, E, Maqueda, M, Cruz, V, Albert, A. | | Deposit date: | 2002-11-25 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Bacteriocin as-48: From Soluble State to Membrane Bound State
J.Mol.Biol., 334, 2003
|
|
1E68
 
 | | Solution structure of bacteriocin AS-48 | | Descriptor: | AS-48 PROTEIN | | Authors: | Gonzalez, C, Langdon, G, Bruix, M, Galvez, A, Valdivia, E, Maqueda, M, Rico, M. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriocin as-48, a Microbial Cyclic Polypeptide Structurally and Functionally Related to Mammalian Nk-Lysin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
