2KMY
 
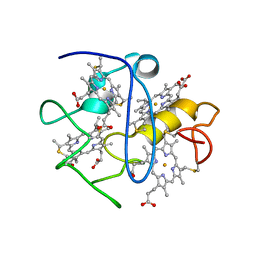 | |
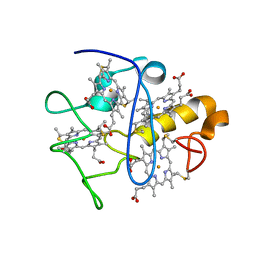
2KSU
 
 | |
2LGN
 
 | | Lactococcin 972 | | Descriptor: | Lactococcin 972 | | Authors: | Turner, D.L, Lamosa, P, Martinez, B. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and properties of lactococcin 972 from Lactococcus lactis
To be Published
|
|
2MS3
 
 | |
2MXK
 
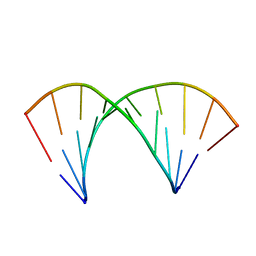 | |
2MXL
 
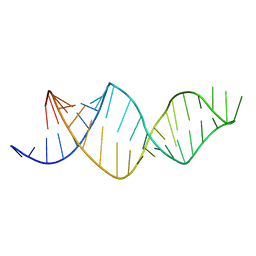 | |
2MXJ
 
 | |
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1QN0
 
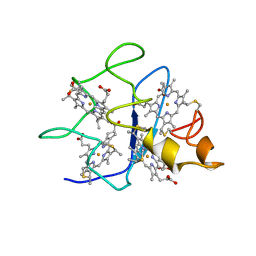 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QN1
 
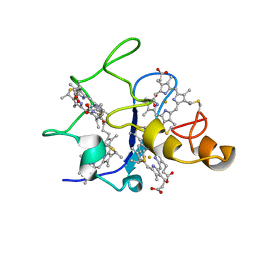 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
333D
 
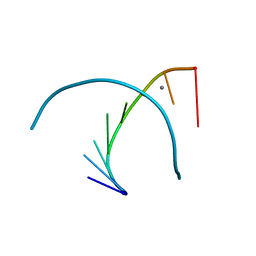 | | THE CRYSTAL STRUCTURE OF AN RNA OLIGOMER INCORPORATING TANDEM ADENOSINE-INOSINE MISMATCHES | | Descriptor: | CALCIUM ION, RNA (5'-R(*CP*GP*CP*AP*IP*GP*CP*G)-3') | | Authors: | Carter, R.J, Baeyens, K.J, SantaLucia Jr, J, Turner, D.H, Holbrook, S.R. | | Deposit date: | 1997-05-20 | | Release date: | 1997-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The crystal structure of an RNA oligomer incorporating tandem adenosine-inosine mismatches.
Nucleic Acids Res., 25, 1997
|
|
1QES
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*UP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
5NEV
 
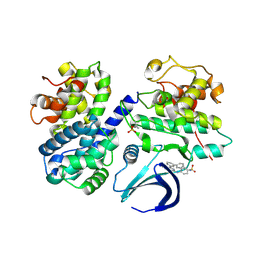 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
5V2R
 
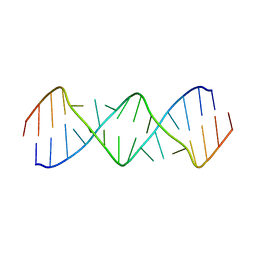 | | Structure of a GA Rich 8x8 Nucleotide RNA Internal Loop | | Descriptor: | RNA (5'-R(*CP*CP*AP*GP*AP*AP*AP*CP*GP*GP*AP*UP*GP*GP*A)-3') | | Authors: | Kauffmann, A.D, Kennedy, S.D, Turner, D.H. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of an 8 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson-Crick Pairs and Comparison to Three-Dimensional Predictions.
Biochemistry, 56, 2017
|
|
2BPN
 
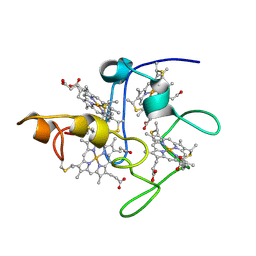 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERRICYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Aguiar, A.P, Brennan, L, Xavier, A.V, Turner, D.L. | | Deposit date: | 2005-04-21 | | Release date: | 2006-03-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Tetrahaem Ferricytochrome C(3) from Desulfovibrio Vulgaris (Hildenborough) and its K45Q Mutant: The Molecular Basis of Cooperativity.
Biochim.Biophys.Acta, 1757, 2006
|
|
1E8E
 
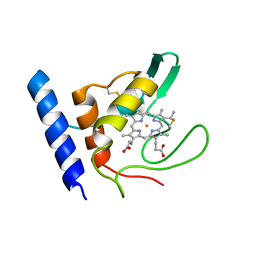 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
1QET
 
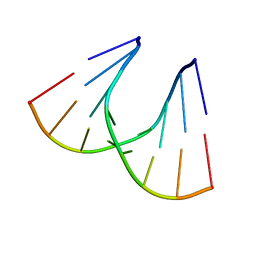 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1F5H
 
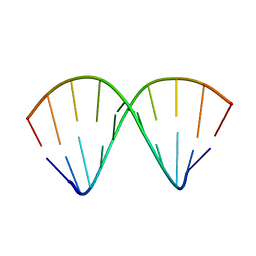 | |
1F5G
 
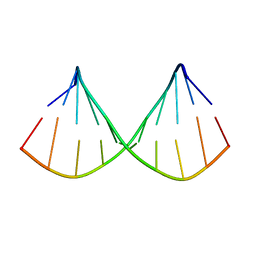 | |
1SPW
 
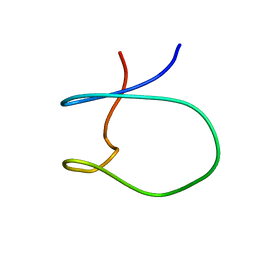 | | Solution Structure of a Loop Truncated Mutant from D. gigas Rubredoxin, NMR | | Descriptor: | Rubredoxin | | Authors: | Pais, T.M, Lamosa, P, dos Santos, W, LeGall, J, Turner, D.L, Santos, H. | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of protein stabilization by solutes: the importance of the hairpin loop in rubredoxins
FEBS J., 272, 2005
|
|
1MV2
 
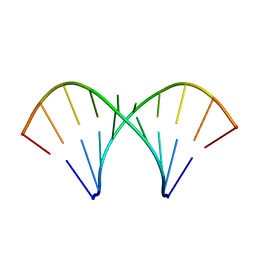 | | The tandem, Face-to-Face AP Pairs in 5'(rGGCAPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MV6
 
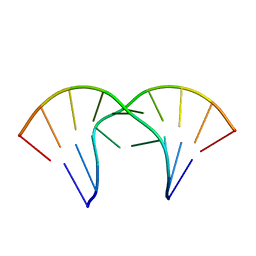 | | The tandem, Sheared PP Pairs in 5'(rGGCPPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1GK5
 
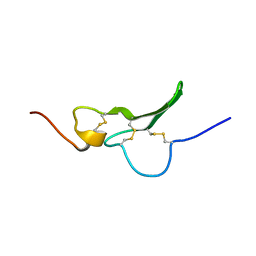 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
1MV1
 
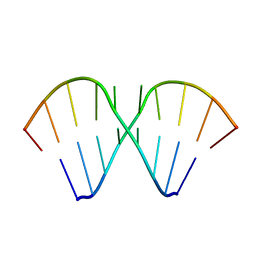 | | The Tandem, Sheared PA Pairs in 5'(rGGCPAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
6N8F
 
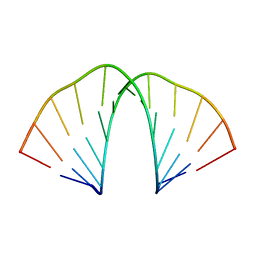 | |
