6ZMV
 
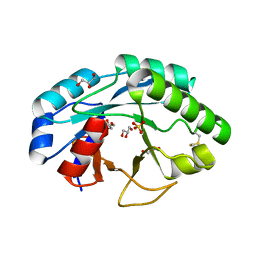 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZM8
 
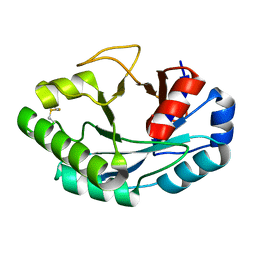 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
7SIE
 
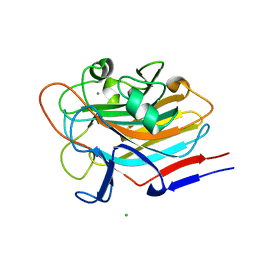 | | Structure of AAP A-domain (residues 351-605) from Staphylococcus epidermidis | | Descriptor: | Accumulation associated protein, CALCIUM ION, CHLORIDE ION | | Authors: | Atkin, K.E, Brentnall, A.S, Dodson, E.J, Whelan, F, Clark, L, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SJK
 
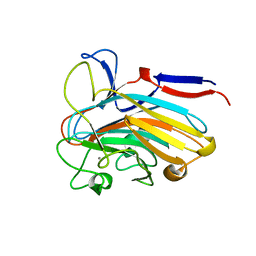 | | Structure of PLS A-domain (residues 391-656) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Pls Plasmin sensitive surface protein | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-18 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SMH
 
 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7SP2
 
 | | Structure of PLS A-domain (residues 391-656; 513-518 deletion mutant) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Plasmin Sensitive Protein Pls | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of PLS A-domain (residues 391-65) from Staphylococcus aureus
Not Published
|
|
4V1R
 
 | | Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, S,R MESO-TARTARIC ACID | | Authors: | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
4TPS
 
 | | Sporulation Inhibitor of DNA Replication, SirA, in complex with Domain I of DnaA | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Chromosomal replication initiator protein DnaA, ... | | Authors: | Jameson, K.H, Turkenburg, J.P, Fogg, M.J, Grahl, A, Wilkinson, A.J. | | Deposit date: | 2014-06-09 | | Release date: | 2014-07-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA.
Mol.Microbiol., 93, 2014
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
4UNE
 
 | | Human insulin B26Phe mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UNG
 
 | | Human insulin B26Asn mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UNH
 
 | | Human insulin B26Gly mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TIP
 
 | |
3TIQ
 
 | | Crystal structure of Staphylococcus aureus SasG G51-E-G52 module | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Surface protein G | | Authors: | Gruszka, D.T, Wojdyla, J.A, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8739 Å) | | Cite: | Staphylococcal biofilm-forming protein has a contiguous rod-like structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZS2
 
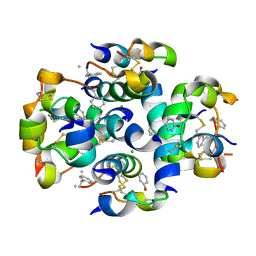 | | TyrB25,NMePheB26,LysB28,ProB29-insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
1O8U
 
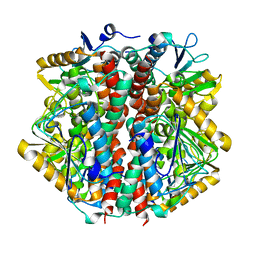 | | The 2 Angstrom Structure of 6-Oxo Camphor Hydrolase: New Structural Diversity in the Crotonase Superfamily | | Descriptor: | 6-OXO CAMPHOR HYDROLASE, SODIUM ION | | Authors: | Grogan, G, Whittingham, J.L, Turkenburg, J.P, Verma, C.S, Walsh, M.A. | | Deposit date: | 2002-12-04 | | Release date: | 2003-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2 a Crystal Structure of 6-Oxo Camphor Hydrolase: New Structural Diversity in the Crotonase Superfamily
J.Biol.Chem., 278, 2003
|
|
1ODT
 
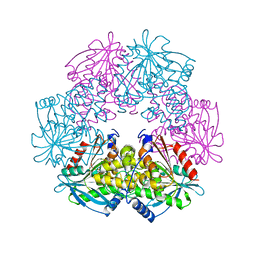 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1ODS
 
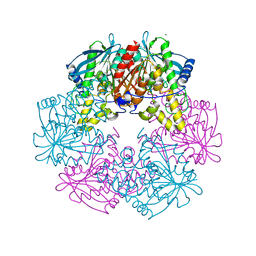 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FJJ
 
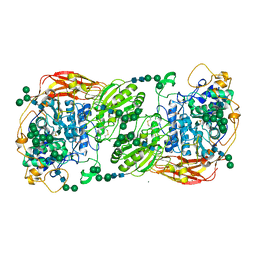 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5G1W
 
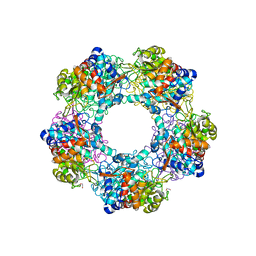 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1U
 
 | | Linalool Dehydratase Isomerase in complex with Geraniol | | Descriptor: | Geraniol, LINALOOL DEHYDRATASE/ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1V
 
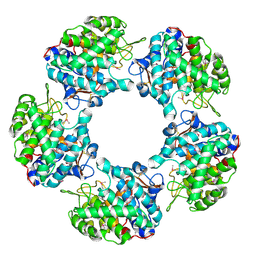 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5LWH
 
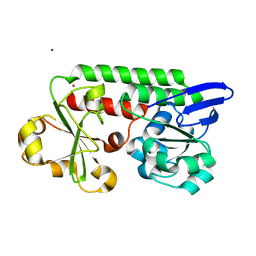 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
