2QDM
 
 | |
3M48
 
 | | GCN4 Leucine Zipper Peptide Mutant | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Du, S, Kettering, R.D, Alvarado, J.J, Tortajada, A, Yeh, J.I. | | Deposit date: | 2010-03-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | GCN4 Leucine Zipper Peptide Mutant
To be Published
|
|
2QDP
 
 | |
2QDC
 
 | |
3D42
 
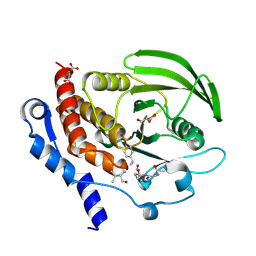 | | Crystal structure of HePTP in complex with a monophosphorylated Erk2 peptide | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Mitogen-activated protein kinase 1 peptide, ... | | Authors: | Critton, D.A, Tortajada, A, Page, R. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of substrate recognition by hematopoietic tyrosine phosphatase.
Biochemistry, 47, 2008
|
|
3D44
 
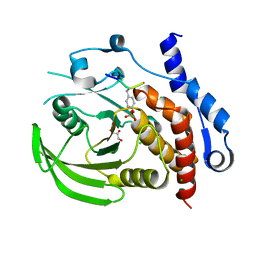 | | Crystal structure of HePTP in complex with a dually phosphorylated Erk2 peptide mimetic | | Descriptor: | CHLORIDE ION, GLYCEROL, Mitogen-activated protein kinase 1 peptide, ... | | Authors: | Critton, D.A, Tortajada, A, Page, R. | | Deposit date: | 2008-05-13 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of substrate recognition by hematopoietic tyrosine phosphatase.
Biochemistry, 47, 2008
|
|
5FR8
 
 | |
5FP2
 
 | |
5FP1
 
 | |
3I1G
 
 | |
