3IVD
 
 | | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine | | Descriptor: | CHLORIDE ION, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine
To be Published
|
|
3IP1
 
 | | Structure of putative alcohol dehydrogenase (TM_042) from Thermotoga maritima | | Descriptor: | Alcohol dehydrogenase, zinc-containing, CADMIUM ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-15 | | Release date: | 2009-09-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of putative alcohol dehydrogenase (TM_042) from Thermotoga maritima
To be published
|
|
3I4F
 
 | | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Ramagopal, U.A, Kim, J, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis
To be published
|
|
3I47
 
 | | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Enoyl CoA hydratase/isomerase (Crotonase) | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila
subsp. pneumophila str. Philadelphia 1
To be Published
|
|
3HU5
 
 | | CRYSTAL STRUCTURE OF isochorismatase family protein from Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | | Descriptor: | Isochorismatase family protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-12 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF isochorismatase family protein from Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough
To be Published
|
|
3HUL
 
 | |
3G1T
 
 | | CRYSTAL STRUCTURE OF short chain dehydrogenase from Salmonella enterica subsp. enterica serovar Typhi str. CT18 | | Descriptor: | MAGNESIUM ION, short chain dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF short chain dehydrogenase from Salmonella enterica subsp. enterica
serovar Typhi str. CT18
To be Published
|
|
3I0T
 
 | | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans | | Descriptor: | BH3703 protein, SULFATE ION | | Authors: | Ramagopal, U.A, Toro, R, Wasserman, S, Burley, S.K, Almo, S.C. | | Deposit date: | 2009-06-25 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans
To be published
|
|
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
3IVE
 
 | | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine
To be Published
|
|
3JZV
 
 | |
5ESR
 
 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
3K4I
 
 | | CRYSTAL STRUCTURE OF uncharacterized protein PSPTO_3204 from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | CRYSTAL STRUCTURE OF uncharacterized protein PSPTO_3204 from Pseudomonas syringae pv. tomato
str. DC3000
To be Published
|
|
3GZ4
 
 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
3H12
 
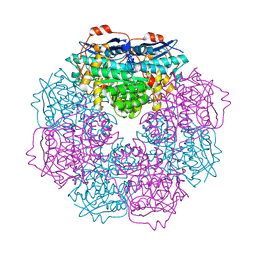 | | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50
To be Published
|
|
3HG9
 
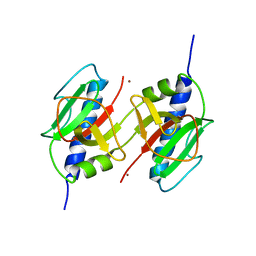 | | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192 | | Descriptor: | NICKEL (II) ION, PilM | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192
To be Published
|
|
3K12
 
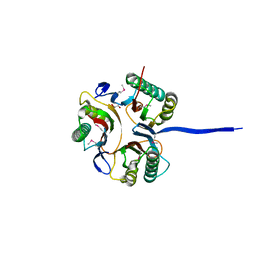 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
3G8R
 
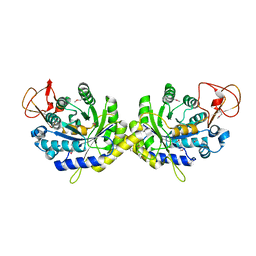 | | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472 | | Descriptor: | Probable spore coat polysaccharide biosynthesis protein E, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472
To be Published
|
|
3JYG
 
 | |
4DHD
 
 | | Crystal structure of isoprenoid synthase A3MSH1 (TARGET EFI-501992) from Pyrobaculum calidifontis | | Descriptor: | ACETATE ION, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4D8L
 
 | | Crystal structure of the 2-pyrone-4,6-dicarboxylic acid hydrolase from sphingomonas paucimobilis | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DI9
 
 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 6.5 | | Descriptor: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ACETATE ION | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DIA
 
 | | CRYSTAL STRUCTURE OF THE D248N mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 4.6 | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4ECJ
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
