3MGK
 
 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3MF4
 
 | | Crystal structure of putative two-component system response regulator/ggdef domain protein | | Descriptor: | MAGNESIUM ION, Two-component system response regulator/GGDEF domain protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative two-component system response regulator/ggdef domain protein
To be Published
|
|
3M0G
 
 | | CRYSTAL STRUCTURE OF putative farnesyl diphosphate synthase from Rhodobacter capsulatus | | Descriptor: | Farnesyl diphosphate synthase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF putative farnesyl diphosphate synthase from Rhodobacter capsulatus
To be Published
|
|
3MWC
 
 | | Crystal structure of probable o-succinylbenzoic acid synthetase from kosmotoga olearia | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-05 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of O-Succinylbenzoic Acid Synthetase from Kosmotoga Olearia
To be Published
|
|
3N28
 
 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | Descriptor: | Phosphoserine phosphatase, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
3MZN
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 | | Descriptor: | ACETATE ION, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3N05
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM STREPTOMYCES AVERMITILIS | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Nh3-Dependent Nad+ Synthetase from Streptomyces Avermitilis
To be Published
|
|
3N4E
 
 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans Pd1222 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans
Pd1222
To be Published
|
|
3N1U
 
 | | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila | | Descriptor: | CALCIUM ION, Hydrolase, HAD superfamily, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila
To be published
|
|
3MY9
 
 | | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Quartararo, C.E, Ramagopal, U, Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans
To be Published
|
|
3NFU
 
 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 complexed with magnesium | | Descriptor: | GLYCEROL, Glucarate dehydratase, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3N4F
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus sp. Y412MC10 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus
sp. Y412MC10
To be Published
|
|
3DP7
 
 | | CRYSTAL STRUCTURE OF SAM-dependent methyltransferase from Bacteroides vulgatus ATCC 8482 | | Descriptor: | SAM-dependent methyltransferase | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | CRYSTAL STRUCTURE OF SAM-dependent methyltransferase from Bacteroides vulgatus ATCC 8482
To be Published
|
|
3N3D
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
3N6J
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z | | Descriptor: | Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z
To be Published
|
|
3FN9
 
 | |
3N6H
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z complexed with magnesium/sulfate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z complexed with magnesium/sulfate
To be Published
|
|
3EWB
 
 | | Crystal structure of N-terminal domain of putative 2-isopropylmalate synthase from Listeria monocytogenes | | Descriptor: | 2-isopropylmalate synthase | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Hu, S, Maletic, M, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of N-terminal domain of putative 2-isopropylmalate synthase from Listeria monocytogenes.
To be Published
|
|
3KTS
 
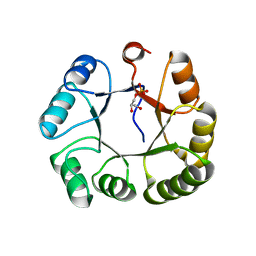 | | CRYSTAL STRUCTURE OF GLYCEROL UPTAKE OPERON ANTITERMINATOR REGULATORY PROTEIN FROM LISTERIA MONOCYTOGENES STR. 4b F2365 | | Descriptor: | Glycerol uptake operon antiterminator regulatory protein, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CRYSTAL STRUCTURE OF GLYCEROL UPTAKE OPERON ANTITERMINATOR REGULATORY PROTEIN FROM LISTERIA MONOCYTOGENES STR. 4b F2365
To be Published
|
|
3MDK
 
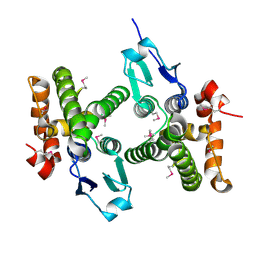 | |
3F5S
 
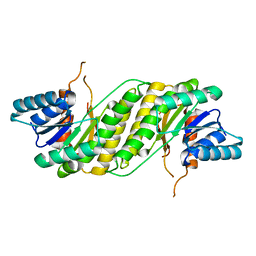 | | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301
To be Published
|
|
3MOG
 
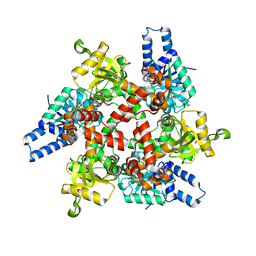 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3M9L
 
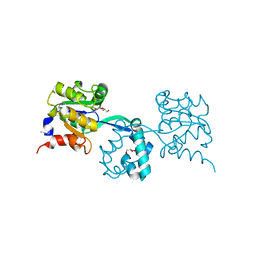 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 | | Descriptor: | GLYCEROL, Hydrolase, haloacid dehalogenase-like family | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Had Family Hydrolase from Pseudomonas Fluorescens Pf-5
To be Published
|
|
3M9U
 
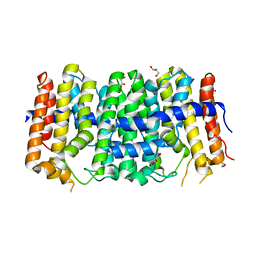 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Farnesyl-diphosphate synthase, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-22 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis Atcc 367
To be Published
|
|
3MDN
 
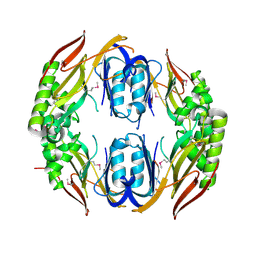 | |
