3ZC9
 
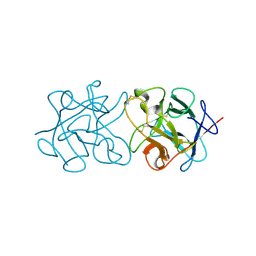 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 4.6 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
5XCH
 
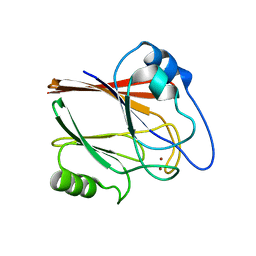 | | Crystal structure of Wild type Vps29 complexed with Zn+2 from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
5XCE
 
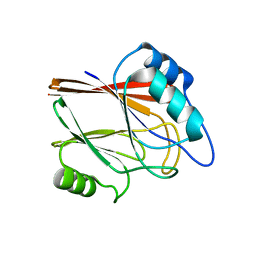 | | Crystal structure of Wild type Vps29 from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
5XCK
 
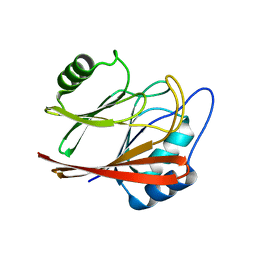 | | Crystal structure of Vps29 double mutant (D62A/H86A) from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
5XCJ
 
 | | Crystal structure of Vps29 single mutant (D62A) from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
3ZC8
 
 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 7.0 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
